Cancer-associated fibroblasts facilitate breast cancer progression through exosomal circTBPL1-mediated intercellular communication
- PMID: 37495592
- PMCID: PMC10372047
- DOI: 10.1038/s41419-023-05986-8
Cancer-associated fibroblasts facilitate breast cancer progression through exosomal circTBPL1-mediated intercellular communication
Abstract
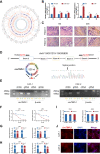
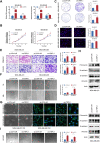
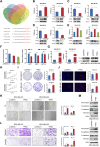
Breast cancer is the major common malignancy worldwide among women. Previous studies reported that cancer-associated fibroblasts (CAFs) showed pivotal roles in regulating tumor progression via exosome-mediated cellular communication. However, the detailed mechanism underlying the exosomal circRNA from CAFs in breast cancer progression remains ambiguous. Here, exosomal circRNA profiling of breast cancer-derived CAFs and normal fibroblasts (NFs) was detected by high-throughput sequencing, and upregulated circTBPL1 expression was identified in CAF exosomes. The exosomal circTBPL1 from CAFs could be transferred to breast cancer cells and promoted cell proliferation, migration, and invasion. Consistently, circTBPL1 knockdown in CAFs attenuated their tumor-promoting ability. Further exploration identified miR-653-5p as an inhibitory target of circTBPL1, and ectopic expression of miR-653-5p could partially reverse the malignant phenotypes induced by circTBPL1 overexpression in breast cancer. Additionally, TPBG was selected as a downstream target gene, and circTBPL1 could protect TPBG from miR-653-5p-mediated degradation, leading to enhanced breast cancer progression. Significantly, the accelerated tumor progression triggered by exosomal circTBPL1 from CAFs was confirmed in xenograft models. Taken together, these results revealed that exosomal circTBPL1 derived from CAFs contributed to cancer progression via miR-653-5p/TPBG pathway, indicating the potential of exosomal circTBPL1 as a biomarker and novel therapeutic target for breast cancer.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Exosomal miR-500a-5p derived from cancer-associated fibroblasts promotes breast cancer cell proliferation and metastasis through targeting USP28.Theranostics. 2021 Feb 6;11(8):3932-3947. doi: 10.7150/thno.53412. eCollection 2021. Theranostics. 2021. PMID: 33664871 Free PMC article.
-
Exosomal miR-146b-5p derived from cancer-associated fibroblasts promotes progression of oral squamous cell carcinoma by downregulating HIPK3.Cell Signal. 2023 Jun;106:110635. doi: 10.1016/j.cellsig.2023.110635. Epub 2023 Feb 20. Cell Signal. 2023. PMID: 36813147
-
Cancer-associated fibroblasts-derived exosome-mediated transfer of miR-345-5p promotes the progression of colorectal cancer by targeting CDKN1A.Carcinogenesis. 2023 Jun 24;44(4):317-327. doi: 10.1093/carcin/bgad014. Carcinogenesis. 2023. PMID: 37052230
-
Exosomal miRNAs and miRNA dysregulation in cancer-associated fibroblasts.Mol Cancer. 2017 Aug 29;16(1):148. doi: 10.1186/s12943-017-0718-4. Mol Cancer. 2017. PMID: 28851377 Free PMC article. Review.
-
The role of CAF derived exosomal microRNAs in the tumour microenvironment of melanoma.Biochim Biophys Acta Rev Cancer. 2021 Jan;1875(1):188456. doi: 10.1016/j.bbcan.2020.188456. Epub 2020 Oct 22. Biochim Biophys Acta Rev Cancer. 2021. PMID: 33153973 Review.
Cited by
-
Recent Advances of Small Extracellular Vesicles for the Regulation and Function of Cancer-Associated Fibroblasts.Int J Mol Sci. 2024 Nov 22;25(23):12548. doi: 10.3390/ijms252312548. Int J Mol Sci. 2024. PMID: 39684264 Free PMC article. Review.
-
Cancer-associated fibroblasts as therapeutic targets for cancer: advances, challenges, and future prospects.J Biomed Sci. 2025 Jan 9;32(1):7. doi: 10.1186/s12929-024-01099-2. J Biomed Sci. 2025. PMID: 39780187 Free PMC article. Review.
-
The role of exosomal non-coding RNAs in the breast cancer tumor microenvironment.Funct Integr Genomics. 2025 Feb 1;25(1):32. doi: 10.1007/s10142-025-01531-2. Funct Integr Genomics. 2025. PMID: 39891771 Review.
-
Cancer-associated fibroblasts affect breast cancer cell sensitivity to chemotherapeutic agents by regulating NRBP2.Toxicol Res (Camb). 2024 Dec 8;13(6):tfae204. doi: 10.1093/toxres/tfae204. eCollection 2024 Dec. Toxicol Res (Camb). 2024. PMID: 39664500
-
Breast cancer shares many epidemiological, lifestyle, and local hormonal and metabolic underpinnings with endometrial and ovarian cancer: a narrative review.Transl Breast Cancer Res. 2025 Jan 21;6:8. doi: 10.21037/tbcr-24-39. eCollection 2025. Transl Breast Cancer Res. 2025. PMID: 39980815 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

