Evolutionary histories of breast cancer and related clones
- PMID: 37495687
- PMCID: PMC10432280
- DOI: 10.1038/s41586-023-06333-9
Evolutionary histories of breast cancer and related clones
Abstract
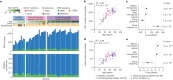
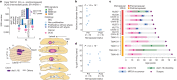
Recent studies have documented frequent evolution of clones carrying common cancer mutations in apparently normal tissues, which are implicated in cancer development1-3. However, our knowledge is still missing with regard to what additional driver events take place in what order, before one or more of these clones in normal tissues ultimately evolve to cancer. Here, using phylogenetic analyses of multiple microdissected samples from both cancer and non-cancer lesions, we show unique evolutionary histories of breast cancers harbouring der(1;16), a common driver alteration found in roughly 20% of breast cancers. The approximate timing of early evolutionary events was estimated from the mutation rate measured in normal epithelial cells. In der(1;16)(+) cancers, the derivative chromosome was acquired from early puberty to late adolescence, followed by the emergence of a common ancestor by the patient's early 30s, from which both cancer and non-cancer clones evolved. Replacing the pre-existing mammary epithelium in the following years, these clones occupied a large area within the premenopausal breast tissues by the time of cancer diagnosis. Evolution of multiple independent cancer founders from the non-cancer ancestors was common, contributing to intratumour heterogeneity. The number of driver events did not correlate with histology, suggesting the role of local microenvironments and/or epigenetic driver events. A similar evolutionary pattern was also observed in another case evolving from an AKT1-mutated founder. Taken together, our findings provide new insight into how breast cancer evolves.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures












References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

