Extensive pedigrees reveal the social organization of a Neolithic community
- PMID: 37495691
- PMCID: PMC10432279
- DOI: 10.1038/s41586-023-06350-8
Extensive pedigrees reveal the social organization of a Neolithic community
Abstract
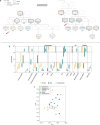
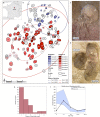
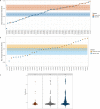
Social anthropology and ethnographic studies have described kinship systems and networks of contact and exchange in extant populations1-4. However, for prehistoric societies, these systems can be studied only indirectly from biological and cultural remains. Stable isotope data, sex and age at death can provide insights into the demographic structure of a burial community and identify local versus non-local childhood signatures, archaeogenetic data can reconstruct the biological relationships between individuals, which enables the reconstruction of pedigrees, and combined evidence informs on kinship practices and residence patterns in prehistoric societies. Here we report ancient DNA, strontium isotope and contextual data from more than 100 individuals from the site Gurgy 'les Noisats' (France), dated to the western European Neolithic around 4850-4500 BC. We find that this burial community was genetically connected by two main pedigrees, spanning seven generations, that were patrilocal and patrilineal, with evidence for female exogamy and exchange with genetically close neighbouring groups. The microdemographic structure of individuals linked and unlinked to the pedigrees reveals additional information about the social structure, living conditions and site occupation. The absence of half-siblings and the high number of adult full siblings suggest that there were stable health conditions and a supportive social network, facilitating high fertility and low mortality5. Age-structure differences and strontium isotope results by generation indicate that the site was used for just a few decades, providing new insights into shifting sedentary farming practices during the European Neolithic.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures








References
-
- Fox, R. Kinship and Marriage: An Anthropological Perspective (Cambridge Univ. Press, 1983).
-
- Morgan, L. H. Ancient Society: Or, Researches in the Lines of Human progress from Savagery, Through Barbarism to Civilization (C. H. Kerr, 1877).
-
- Lévi-Strauss, C. The Elementary Structures of Kinship (Beacon Press, 1971).
-
- Schneider, D. M. A Critique of the Study of Kinship (Univ. Michigan Press, 1984).
-
- Bocquet-Appel J-P. Paleoanthropological traces of a Neolithic demographic transition. Curr. Anthropol. 2002;43:637–650. doi: 10.1086/342429. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

