Isolation of a bacteriophage targeting Pseudomonas aeruginosa and exhibits a promising in vivo efficacy
- PMID: 37495819
- PMCID: PMC10371947
- DOI: 10.1186/s13568-023-01582-3
Isolation of a bacteriophage targeting Pseudomonas aeruginosa and exhibits a promising in vivo efficacy
Abstract
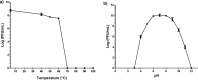
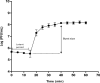
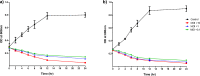
Pseudomonas aeruginosa is an important pathogen that causes serious infections. Bacterial biofilms are highly resistant and render bacterial treatment very difficult, therefore necessitates alternative antibacterial strategies. Phage therapy has been recently regarded as a potential therapeutic option for treatment of bacterial infections. In the current study, a novel podovirus vB_PaeP_PS28 has been isolated from sewage with higher lytic activity against P. aeruginosa. Isolated phage exhibits a short latent period, large burst size and higher stability over a wide range of temperatures and pH. The genome of vB_PaeP_PS28 consists of 72,283 bp circular double-stranded DNA, with G + C content of 54.75%. The phage genome contains 94 open reading frames (ORFs); 32 for known functional proteins and 62 for hypothetical proteins and no tRNA genes. The phage vB_PaeP_PS28 effectively inhibited the growth of P. aeruginosa planktonic cells and displayed a higher biofilm degrading capability. Moreover, therapeutic efficacy of isolated phage was evaluated in vivo using mice infection model. Interestingly, survival of mice infected with P. aeruginosa was significantly enhanced upon treatment with vB_PaeP_PS28. Furthermore, the bacterial load in liver and kidney isolated from mice infected with P. aeruginosa and treated with phage markedly decreased as compared with phage-untreated P. aeruginosa-infected mice. These findings support the efficacy of isolated phage vB_PaeP_PS28 in reducing P. aeruginosa colonization and pathogenesis in host. Importantly, the isolated phage vB_PaeP_PS28 could be applied alone or as combination therapy with other lytic phages as phage cocktail therapy or with antibiotics to limit infections caused by P. aeruginosa.
Keywords: Antibiotic resistance; Bacteriophage; Biofilm; Genomic analysis; Phage therapy; Pseudomonas aeruginosa.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







Similar articles
-
vB_PaeP_PZH3, a novel bacteriophage for the treatment of MDR Pseudomonas aeruginosa in a mouse wound infection model.J Glob Antimicrob Resist. 2025 Jun 28;44:197-206. doi: 10.1016/j.jgar.2025.06.019. Online ahead of print. J Glob Antimicrob Resist. 2025. PMID: 40588032
-
A novel lytic phage exhibiting a remarkable in vivo therapeutic potential and higher antibiofilm activity against Pseudomonas aeruginosa.Eur J Clin Microbiol Infect Dis. 2023 Oct;42(10):1207-1234. doi: 10.1007/s10096-023-04649-y. Epub 2023 Aug 23. Eur J Clin Microbiol Infect Dis. 2023. PMID: 37608144 Free PMC article.
-
Characterization of Pseudomonas aeruginosa bacteriophages and control hemorrhagic pneumonia on a mice model.Front Microbiol. 2024 May 14;15:1396774. doi: 10.3389/fmicb.2024.1396774. eCollection 2024. Front Microbiol. 2024. PMID: 38808279 Free PMC article.
-
Isolation and Characterization of a Novel Lytic Phage, vB_PseuP-SA22, and Its Efficacy against Carbapenem-Resistant Pseudomonas aeruginosa.Antibiotics (Basel). 2023 Mar 2;12(3):497. doi: 10.3390/antibiotics12030497. Antibiotics (Basel). 2023. Retraction in: Antibiotics (Basel). 2024 Aug 28;13(9):814. doi: 10.3390/antibiotics13090814. PMID: 36978364 Free PMC article. Retracted.
-
Characterization of a lytic Pseudomonas aeruginosa phage vB_PaeP_ASP23 and functional analysis of its lysin LysASP and holin HolASP.Front Microbiol. 2023 Mar 15;14:1093668. doi: 10.3389/fmicb.2023.1093668. eCollection 2023. Front Microbiol. 2023. PMID: 36998407 Free PMC article.
Cited by
-
Molecular Aspects of the Functioning of Pathogenic Bacteria Biofilm Based on Quorum Sensing (QS) Signal-Response System and Innovative Non-Antibiotic Strategies for Their Elimination.Int J Mol Sci. 2024 Feb 24;25(5):2655. doi: 10.3390/ijms25052655. Int J Mol Sci. 2024. PMID: 38473900 Free PMC article. Review.
-
Miconazole and phenothiazine hinder the quorum sensing regulated virulence in Pseudomonas aeruginosa.J Antibiot (Tokyo). 2024 Jul;77(7):454-465. doi: 10.1038/s41429-024-00731-5. Epub 2024 May 9. J Antibiot (Tokyo). 2024. PMID: 38724627 Free PMC article.
-
Analysis of a novel phage as a promising biological agent targeting multidrug resistant Klebsiella pneumoniae.AMB Express. 2025 Mar 5;15(1):37. doi: 10.1186/s13568-025-01846-0. AMB Express. 2025. PMID: 40044971 Free PMC article.
-
The Potential of Phage Treatment to Inactivate Planktonic and Biofilm-Forming Pseudomonas aeruginosa.Microorganisms. 2024 Aug 29;12(9):1795. doi: 10.3390/microorganisms12091795. Microorganisms. 2024. PMID: 39338470 Free PMC article.
-
Isolation of Streptococcus mutans temperate bacteriophage with broad killing activity to S. mutans clinical isolates.iScience. 2023 Nov 14;26(12):108465. doi: 10.1016/j.isci.2023.108465. eCollection 2023 Dec 15. iScience. 2023. PMID: 38089578 Free PMC article.
References
-
- Ackermann HW, Tremblay D, Moineau S. Long-term bacteriophage preservation. WFCC Newsl. 2004;38:35–40.
-
- Adnan M, Shah MRA, Jamal M, Jalil F, Andleeb S, Nawaz MA, Pervez S, Hussain T, Shah I, Imran M. Isolation and characterization of bacteriophage to control multidrug-resistant Pseudomonas aeruginosa planktonic cells and biofilm. Biologicals. 2020;63:89–96. doi: 10.1016/j.biologicals.2019.10.003. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

