Integrated genetic analyses of immunodeficiency-associated Epstein-Barr virus- (EBV) positive primary CNS lymphomas
- PMID: 37495858
- PMCID: PMC10412493
- DOI: 10.1007/s00401-023-02613-w
Integrated genetic analyses of immunodeficiency-associated Epstein-Barr virus- (EBV) positive primary CNS lymphomas
Abstract
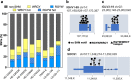
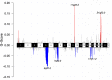
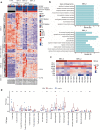
Immunodeficiency-associated primary CNS lymphoma (PCNSL) represents a distinct clinicopathological entity, which is typically Epstein-Barr virus-positive (EBV+) and carries an inferior prognosis. Genetic alterations that characterize EBV-related CNS lymphomagenesis remain unclear precluding molecular classification and targeted therapies. In this study, a comprehensive genetic analysis of 22 EBV+ PCNSL, therefore, integrated clinical and pathological information with exome and RNA sequencing (RNASeq) data. EBV+ PCNSL with germline controls carried a median of 55 protein-coding single nucleotide variants (SNVs; range 24-217) and 2 insertions/deletions (range 0-22). Genetic landscape was largely shaped by aberrant somatic hypermutation with a median of 41.01% (range 31.79-53.49%) of SNVs mapping to its target motifs. Tumors lacked established SNVs (MYD88, CD79B, PIM1) and copy number variants (CDKN2A, HLA loss) driving EBV- PCNSL. Instead, EBV+ PCNSL were characterized by SOCS1 mutations (26%), predicted to disinhibit JAK/STAT signaling, and mutually exclusive gain-of-function NOTCH pathway SNVs (26%). Copy number gains were enriched on 11q23.3, a locus directly targeted for chromosomal aberrations by EBV, that includes SIK3 known to protect from cytotoxic T-cell responses. Losses covered 5q31.2 (STING), critical for sensing viral DNA, and 17q11 (NF1). Unsupervised clustering of RNASeq data revealed two distinct transcriptional groups, that shared strong expression of CD70 and IL1R2, previously linked to tolerogenic tumor microenvironments. Correspondingly, deconvolution of bulk RNASeq data revealed elevated M2-macrophage, T-regulatory cell, mast cell and monocyte fractions in EBV+ PCNSL. In addition to novel insights into the pathobiology of EBV+ PCNSL, the data provide the rationale for the exploration of targeted therapies including JAK-, NOTCH- and CD70-directed approaches.
Keywords: Epstein-Barr virus; Exome sequencing; Genetics; Immunodeficiency; Non-Hodgkin lymphoma; Primary CNS lymphoma; RNA sequencing.
© 2023. The Author(s).
Conflict of interest statement
Dr. Kaulen reports no disclosures. Dr. Denisova reports no disclosures. Dr. Hinz reports no disclosures. Ms. Hai reports no disclosures. Mr. Friedel reports no disclosures. Dr. Henegariu reports no disclosures. Mr. Hoffmann reports no disclosures. Mr. Ito reports no disclosures. Mr. Kourtesakis reports no disclosures. Mr. Lehnert reports no disclosures. Dr. Doubrovinskaia reports no disclosures. Dr. Karschnia reports no disclosures. Dr. von Baumgarten reports no disclosures. Dr. Kessler reports no disclosures. Dr. Baehring reports no disclosures. Dr. Brors reports no disclosures. Dr. Sahm reports honoraria from Illumina, Agilent and Bayer, patents on methylation-based classification managed by DKFZ, and being cofounder and shareholder of Heidelberg Epignostix GmbH. Dr. Wick reports support for clinical trials from Apogenix, Pfizer, Roche and Vaximm as well as honoraria for advice from Astra Zeneca, Enterome, MSD, Roche, Servier and Vaximm paid to the Medical Faculty.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

