Inhibitors of riboflavin biosynthetic pathway enzymes as potential antibacterial drugs
- PMID: 37496776
- PMCID: PMC10366380
- DOI: 10.3389/fmolb.2023.1228763
Inhibitors of riboflavin biosynthetic pathway enzymes as potential antibacterial drugs
Abstract
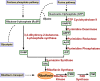
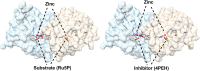
Multiple drug resistance is the main obstacle in the treatment of bacterial diseases. Resistance against antibiotics demands the exploration of new antimicrobial drug targets. A variety of in silico and genetic approaches show that the enzymes of the riboflavin biosynthetic pathway are crucial for the survival of bacteria. This pathway is absent in humans thus enzymes of the riboflavin biosynthetic pathway are emerging drug targets for resistant pathogenic bacterial strains. Exploring the structural details, their mechanism of action, intermediate elucidation, and interaction analysis would help in designing suitable inhibitors of these enzymes. The riboflavin biosynthetic pathway consists of seven distinct enzymes, namely, 3,4-dihydroxy-2-butanone 4-phosphate synthase, GTP cyclohydrolase II, pyrimidine deaminase/reductase, phosphatase, lumazine synthase, and riboflavin synthase. The present review summarizes the research work that has been carried out on these enzymes in terms of their structures, active site architectures, and molecular mechanism of catalysis. This review also walks through small molecule inhibitors that have been developed against several of these enzymes.
Keywords: 3,4-dihydroxy-2-butanone 4-phosphate synthase; inhibitors; lumazine synthase; riboflavin biosynthetic pathway; riboflavin synthase.
Copyright © 2023 Islam and Kumar.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
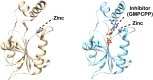

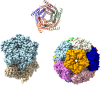
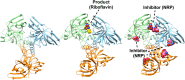
References
Publication types
LinkOut - more resources
Full Text Sources

