Unraveling Impacts of Chamber-Specific Differences in Intercalated Disc Ultrastructure and Molecular Organization on Cardiac Conduction
- PMID: 37498248
- PMCID: PMC11102000
- DOI: 10.1016/j.jacep.2023.05.042
Unraveling Impacts of Chamber-Specific Differences in Intercalated Disc Ultrastructure and Molecular Organization on Cardiac Conduction
Abstract
Background: Propagation of action potentials through the heart coordinates the heartbeat. Thus, intercalated discs, specialized cell-cell contact sites that provide electrical and mechanical coupling between cardiomyocytes, are an important target for study. Impaired propagation leads to arrhythmias in many pathologies, where intercalated disc remodeling is a common finding, hence the importance and urgency of understanding propagation dependence on intercalated disc structure. Conventional modeling approaches cannot predict changes in propagation elicited by perturbations that alter intercalated disc ultrastructure or molecular organization, because of lack of quantitative structural data at subcellular through nano scales.
Objectives: This study sought to quantify intercalated disc structure at these spatial scales in the healthy adult mouse heart and relate them to chamber-specific properties of propagation as a precursor to understanding the effects of pathological intercalated disc remodeling.
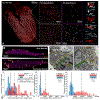
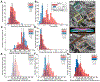
Methods: Using super-resolution light microscopy, electron microscopy, and computational image analysis, we provide here the first ever systematic, multiscale quantification of intercalated disc ultrastructure and molecular organization.
Results: By incorporating these data into a rule-based model of cardiac tissue with realistic intercalated disc structure, and comparing model predictions of electrical propagation with experimental measures of conduction velocity, we reveal that atrial intercalated discs can support faster conduction than their ventricular counterparts, which is normally masked by interchamber differences in myocyte geometry. Further, we identify key ultrastructural and molecular organization features underpinning the ability of atrial intercalated discs to support faster conduction.
Conclusions: These data provide the first stepping stone to elucidating chamber-specific effects of pathological intercalated disc remodeling, as occurs in many arrhythmic diseases.
Keywords: cardiac conduction; gap junctions; intercalated disc; ion channels; microscopy.
Copyright © 2023 American College of Cardiology Foundation. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Funding Support and Author Disclosures This work was supported by National Institutes of Health R01 grants (HL148736 awarded to Dr Veeraraghavan, HL138003 awarded to Dr Weinberg, and R01HL155378 and R01 NS121234 awarded to Dr Radwański) as well as an American Heart Association grant (20TPA35460040) awarded to Dr Veeraraghavan, an American Heart Association Predoctoral Fellowship awarded to Dr Struckman, an American Heart Association Postdoctoral Fellowship awarded to Dr Moise, and a National Institutes of Health T32 (HL149637-02) and L40 (NS129034) awarded to Dr King. All other authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Figures
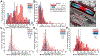


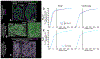

Update of
-
Unraveling Chamber-specific Differences in Intercalated Disc Ultrastructure and Molecular Organization and Their Impact on Cardiac Conduction.bioRxiv [Preprint]. 2023 Mar 24:2023.02.13.528369. doi: 10.1101/2023.02.13.528369. bioRxiv. 2023. Update in: JACC Clin Electrophysiol. 2023 Dec;9(12):2425-2443. doi: 10.1016/j.jacep.2023.05.042. PMID: 36824727 Free PMC article. Updated. Preprint.
Similar articles
-
Unraveling Chamber-specific Differences in Intercalated Disc Ultrastructure and Molecular Organization and Their Impact on Cardiac Conduction.bioRxiv [Preprint]. 2023 Mar 24:2023.02.13.528369. doi: 10.1101/2023.02.13.528369. bioRxiv. 2023. Update in: JACC Clin Electrophysiol. 2023 Dec;9(12):2425-2443. doi: 10.1016/j.jacep.2023.05.042. PMID: 36824727 Free PMC article. Updated. Preprint.
-
Unravelling the ultrastructural details of αT-catenin-deficient cell-cell contacts between heart muscle cells by the use of FIB-SEM.J Microsc. 2020 Sep;279(3):189-196. doi: 10.1111/jmi.12855. Epub 2019 Dec 22. J Microsc. 2020. PMID: 31828778
-
Distribution of cardiac sodium channels in clusters potentiates ephaptic interactions in the intercalated disc.J Physiol. 2018 Feb 15;596(4):563-589. doi: 10.1113/JP275351. Epub 2018 Jan 9. J Physiol. 2018. PMID: 29210458 Free PMC article.
-
Role of gap junctions in the propagation of the cardiac action potential.Cardiovasc Res. 2004 May 1;62(2):309-22. doi: 10.1016/j.cardiores.2003.11.035. Cardiovasc Res. 2004. PMID: 15094351 Review.
-
The intercalated disc: a unique organelle for electromechanical synchrony in cardiomyocytes.Physiol Rev. 2023 Jul 1;103(3):2271-2319. doi: 10.1152/physrev.00021.2022. Epub 2023 Feb 2. Physiol Rev. 2023. PMID: 36731030 Free PMC article. Review.
Cited by
-
Indirect Correlative Light and Electron Microscopy (iCLEM): A Novel Pipeline for Multiscale Quantification of Structure From Molecules to Organs.Microsc Microanal. 2024 Apr 29;30(2):318-333. doi: 10.1093/mam/ozae021. Microsc Microanal. 2024. PMID: 38525890 Free PMC article.
-
Vascular Endothelial Barrier Protection Prevents Atrial Fibrillation by Preserving Cardiac Nanostructure.JACC Clin Electrophysiol. 2023 Dec;9(12):2444-2458. doi: 10.1016/j.jacep.2023.10.013. Epub 2023 Nov 29. JACC Clin Electrophysiol. 2023. PMID: 38032579 Free PMC article.
-
Lamotrigine promotes reentrant ventricular tachycardia in murine hearts.Epilepsia. 2025 May;66(5):1691-1702. doi: 10.1111/epi.18295. Epub 2025 Jan 30. Epilepsia. 2025. PMID: 39887338 Free PMC article.
-
New focus on cardiac voltage-gated sodium channel β1 and β1B: Novel targets for treating and understanding arrhythmias?Heart Rhythm. 2025 Jan;22(1):181-191. doi: 10.1016/j.hrthm.2024.06.029. Epub 2024 Jun 21. Heart Rhythm. 2025. PMID: 38908461 Free PMC article. Review.
-
Amniotic membrane, a novel bioscaffold in cardiac diseases: from mechanism to applications.Front Bioeng Biotechnol. 2024 Dec 20;12:1521462. doi: 10.3389/fbioe.2024.1521462. eCollection 2024. Front Bioeng Biotechnol. 2024. PMID: 39758951 Free PMC article. Review.
References
-
- Kleber AG, Rudy Y. Basic mechanisms of cardiac impulse propagation and associated arrhythmias. Physiol Rev. 2004;84:431–488. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

