IFF: Identifying key residues in intrinsically disordered regions of proteins using machine learning
- PMID: 37498545
- PMCID: PMC10443345
- DOI: 10.1002/pro.4739
IFF: Identifying key residues in intrinsically disordered regions of proteins using machine learning
Abstract
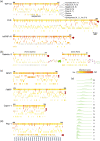
Conserved residues in protein homolog sequence alignments are structurally or functionally important. For intrinsically disordered proteins or proteins with intrinsically disordered regions (IDRs), however, alignment often fails because they lack a steric structure to constrain evolution. Although sequences vary, the physicochemical features of IDRs may be preserved in maintaining function. Therefore, a method to retrieve common IDR features may help identify functionally important residues. We applied unsupervised contrastive learning to train a model with self-attention neuronal networks on human IDR orthologs. Parameters in the model were trained to match sequences in ortholog pairs but not in other IDRs. The trained model successfully identifies previously reported critical residues from experimental studies, especially those with an overall pattern (e.g., multiple aromatic residues or charged blocks) rather than short motifs. This predictive model can be used to identify potentially important residues in other proteins, improving our understanding of their functions. The trained model can be run directly from the Jupyter Notebook in the GitHub repository using Binder (mybinder.org). The only required input is the primary sequence. The training scripts are available on GitHub (https://github.com/allmwh/IFF). The training datasets have been deposited in an Open Science Framework repository (https://osf.io/jk29b).
Keywords: intrinsically disordered proteins; liquid-liquid phase separation; unsupervised contrastive machine learning.
© 2023 The Protein Society.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
SHARK enables sensitive detection of evolutionary homologs and functional analogs in unalignable and disordered sequences.Proc Natl Acad Sci U S A. 2024 Oct 15;121(42):e2401622121. doi: 10.1073/pnas.2401622121. Epub 2024 Oct 9. Proc Natl Acad Sci U S A. 2024. PMID: 39383002 Free PMC article.
-
Discovering molecular features of intrinsically disordered regions by using evolution for contrastive learning.PLoS Comput Biol. 2022 Jun 29;18(6):e1010238. doi: 10.1371/journal.pcbi.1010238. eCollection 2022 Jun. PLoS Comput Biol. 2022. PMID: 35767567 Free PMC article.
-
The return of the rings: Evolutionary convergence of aromatic residues in the intrinsically disordered regions of RNA-binding proteins for liquid-liquid phase separation.Protein Sci. 2022 May;31(5):e4317. doi: 10.1002/pro.4317. Protein Sci. 2022. PMID: 35481633 Free PMC article.
-
Towards Decoding the Sequence-Based Grammar Governing the Functions of Intrinsically Disordered Protein Regions.J Mol Biol. 2021 Jun 11;433(12):166724. doi: 10.1016/j.jmb.2020.11.023. Epub 2020 Nov 26. J Mol Biol. 2021. PMID: 33248138 Review.
-
Features of molecular recognition of intrinsically disordered proteins via coupled folding and binding.Protein Sci. 2019 Nov;28(11):1952-1965. doi: 10.1002/pro.3718. Epub 2019 Sep 4. Protein Sci. 2019. PMID: 31441158 Free PMC article. Review.
Cited by
-
SHARK: web server for alignment-free homology assessment for intrinsically disordered and unalignable protein regions.Nucleic Acids Res. 2025 Jul 7;53(W1):W512-W519. doi: 10.1093/nar/gkaf408. Nucleic Acids Res. 2025. PMID: 40396357 Free PMC article.
-
SHARK enables sensitive detection of evolutionary homologs and functional analogs in unalignable and disordered sequences.Proc Natl Acad Sci U S A. 2024 Oct 15;121(42):e2401622121. doi: 10.1073/pnas.2401622121. Epub 2024 Oct 9. Proc Natl Acad Sci U S A. 2024. PMID: 39383002 Free PMC article.
-
SHARK-capture identifies functional motifs in intrinsically disordered protein regions.Protein Sci. 2025 Apr;34(4):e70091. doi: 10.1002/pro.70091. Protein Sci. 2025. PMID: 40100159 Free PMC article.
References
-
- Alberti S, Hyman AA. Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat Rev Mol Cell Biol. 2021;22(3):196–213. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

