Targeting collagen XVIII improves the efficiency of ErbB inhibitors in breast cancer models
- PMID: 37498672
- PMCID: PMC10702473
- DOI: 10.1172/JCI159181
Targeting collagen XVIII improves the efficiency of ErbB inhibitors in breast cancer models
Abstract
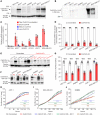
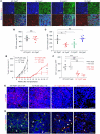
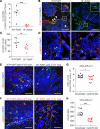
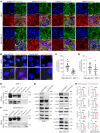
The tumor extracellular matrix (ECM) critically regulates cancer progression and treatment response. Expression of the basement membrane component collagen XVIII (ColXVIII) is induced in solid tumors, but its involvement in tumorigenesis has remained elusive. We show here that ColXVIII was markedly upregulated in human breast cancer (BC) and was closely associated with a poor prognosis in high-grade BCs. We discovered a role for ColXVIII as a modulator of epidermal growth factor receptor tyrosine kinase (ErbB) signaling and show that it forms a complex with ErbB1 and -2 (also known as EGFR and human epidermal growth factor receptor 2 [HER2]) and α6-integrin to promote cancer cell proliferation in a pathway involving its N-terminal portion and the MAPK/ERK1/2 and PI3K/AKT cascades. Studies using Col18a1 mouse models crossed with the mouse mammary tumor virus-polyoma virus middle T antigen (MMTV-PyMT) mammary carcinogenesis model showed that ColXVIII promoted BC growth and metastasis in a tumor cell-autonomous manner. Moreover, the number of mammary cancer stem cells was significantly reduced in the MMTV-PyMT and human cell models upon ColXVIII inhibition. Finally, ablation of ColXVIII substantially improved the efficacy of ErbB-targeting therapies in both preclinical models. In summary, ColXVIII was found to sustain the stemness properties of BC cells and tumor progression and metastasis through ErbB signaling, suggesting that targeting ColXVIII in the tumor milieu may have important therapeutic potential.
Keywords: Breast cancer; Collagens; Growth factors; Oncology.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

