In vivo hematopoietic stem cell modification by mRNA delivery
- PMID: 37499029
- PMCID: PMC10567133
- DOI: 10.1126/science.ade6967
In vivo hematopoietic stem cell modification by mRNA delivery
Erratum in
-
Erratum for the Research Article "In vivo hematopoietic stem cell modification by mRNA delivery" by L. Breda et al.Science. 2025 Jun 5;388(6751):eadz0744. doi: 10.1126/science.adz0744. Epub 2025 Jun 5. Science. 2025. PMID: 40472109 No abstract available.
Abstract
Hematopoietic stem cells (HSCs) are the source of all blood cells over an individual's lifetime. Diseased HSCs can be replaced with gene-engineered or healthy HSCs through HSC transplantation (HSCT). However, current protocols carry major side effects and have limited access. We developed CD117/LNP-messenger RNA (mRNA), a lipid nanoparticle (LNP) that encapsulates mRNA and is targeted to the stem cell factor receptor (CD117) on HSCs. Delivery of the anti-human CD117/LNP-based editing system yielded near-complete correction of hematopoietic sickle cells. Furthermore, in vivo delivery of pro-apoptotic PUMA (p53 up-regulated modulator of apoptosis) mRNA with CD117/LNP affected HSC function and permitted nongenotoxic conditioning for HSCT. The ability to target HSCs in vivo offers a nongenotoxic conditioning regimen for HSCT, and this platform could be the basis of in vivo genome editing to cure genetic disorders, which would abrogate the need for HSCT.
Conflict of interest statement
S.R. is a member of scientific advisory board of Ionis Pharmaceuticals, Meira GTx, Vifor and Disc Medicine. H.P., and D.W. are scientific founders and hold equity in Capstan Therapeutics. Y.K.T. and B.L.M. are employees and hold equity in Acuitas Therapeutics. D.W. and H.P. receive research support from BioNTech. L.B., T.E.P., M.P.T., S.R., and H.P. are inventors (University of Pennsylvania) on a patent for the in vivo gene editing of hematopoietic stem cells using targeted LNP for hemoglobinopathies (US Provisional Patent Application 63/499,580 filed 2 May 2023). L.B., T.E.P., M.P.T., S.R., and H.P. are inventors (University of Pennsylvania) on a patent for HSC-targeted LNP-mRNA for conditioning before HSC transplant (US Provisional Patent Application 046483-6260 filed 9 December 2022). D.W., and H.P. are inventors (University of Pennsylvania) on a patent on the compositions and methods for targeting lipid nanoparticle mRNA therapeutics to stem cells (US Provisional Patent Application 63/182,639 filed 30 April 2021, WIPO Patent Application PCT/US2022/026933). In accordance with the University of Pennsylvania policies and procedures and our ethical obligations as researchers, D.W. and H.P. are named on additional patents that describe the use of nucleoside-modified mRNA and targeted LNPs as platforms to deliver therapeutic proteins and vaccines. These interests have been fully disclosed to the University of Pennsylvania, and approved plans are in place for managing any potential conflicts arising from licensing these patents.
Figures


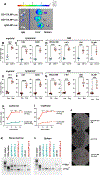

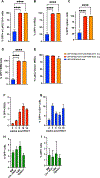
Comment in
-
A step closer to in vivo editing of haematopoietic stem cells.Nat Rev Drug Discov. 2023 Oct;22(10):786. doi: 10.1038/d41573-023-00147-0. Nat Rev Drug Discov. 2023. PMID: 37773516 No abstract available.
References
-
- Kent D et al. , Regulation of hematopoietic stem cells by the steel factor/KIT signaling pathway. Clin Cancer Res 14, 1926–1930 (2008). - PubMed
-
- Yee NS, Hsiau CW, Serve H, Vosseller K, Besmer P, Mechanism of down-regulation of c-kit receptor. Roles of receptor tyrosine kinase, phosphatidylinositol 3’-kinase, and protein kinase C. J Biol Chem 269, 31991–31998 (1994). - PubMed
-
- Kariko K, Buckstein M, Ni H, Weissman D, Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23, 165–175 (2005). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

