The microbial genotoxin colibactin exacerbates mismatch repair mutations in colorectal tumors
- PMID: 37499275
- PMCID: PMC10413156
- DOI: 10.1016/j.neo.2023.100918
The microbial genotoxin colibactin exacerbates mismatch repair mutations in colorectal tumors
Abstract
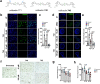
Certain Enterobacteriaceae strains contain a 54-kb biosynthetic gene cluster referred to as "pks" encoding the biosynthesis of a secondary metabolite, colibactin. Colibactin-producing E. coli promote colorectal cancer (CRC) in preclinical models, and in vitro induce a specific mutational signature that is also detected in human CRC genomes. Yet, how colibactin exposure affects the mutational landscape of CRC in vivo remains unclear. Here we show that colibactin-producing E. coli-driven colonic tumors in mice have a significantly higher SBS burden and a larger percentage of these mutations can be attributed to a signature associated with mismatch repair deficiency (MMRd; SBS15), compared to tumors developed in the presence of colibactin-deficient E. coli. We found that the synthetic colibactin 742 but not an inactive analog 746 causes DNA damage and induces transcriptional activation of p53 and senescence signaling pathways in non-transformed human colonic epithelial cells. In MMRd colon cancer cells (HCT 116), chronic exposure to 742 resulted in the upregulation of BRCA1, Fanconi anemia, and MMR signaling pathways as revealed by global transcriptomic analysis. This was accompanied by increased T>N single-base substitutions (SBS) attributed to the proposed pks+E. coli signature (SBS88), reactive oxygen species (SBS17), and mismatch-repair deficiency (SBS44). A significant co-occurrence between MMRd SBS44 and pks-associated SBS88 signature was observed in a large cohort of human CRC patients (n=2,945), and significantly more SBS44 mutations were found when SBS88 was also detected. Collectively, these findings reveal the host response mechanisms underlying colibactin genotoxic activity and suggest that colibactin may exacerbate MMRd-associated mutations.
Keywords: Colibactin; Colorectal cancer; Escherichia coli; Mismatch repair; Mutation.
Copyright © 2023. Published by Elsevier Inc.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
-
- Nougayrède J.P., Homburg S., Taieb F., Boury M., Brzuszkiewicz E., Gottschalk G., et al. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science. 2006;313:848–851. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

