Visualization of direct and diffusion-assisted RAD51 nucleation by full-length human BRCA2 protein
- PMID: 37499663
- PMCID: PMC7615647
- DOI: 10.1016/j.molcel.2023.06.031
Visualization of direct and diffusion-assisted RAD51 nucleation by full-length human BRCA2 protein
Abstract
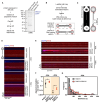
Homologous recombination (HR) is essential for error-free repair of DNA double-strand breaks, perturbed replication forks (RFs), and post-replicative single-stranded DNA (ssDNA) gaps. To initiate HR, the recombination mediator and tumor suppressor protein BRCA2 facilitates nucleation of RAD51 on ssDNA prior to stimulation of RAD51 filament growth by RAD51 paralogs. Although ssDNA binding by BRCA2 has been implicated in RAD51 nucleation, the function of double-stranded DNA (dsDNA) binding by BRCA2 remains unclear. Here, we exploit single-molecule (SM) imaging to visualize BRCA2-mediated RAD51 nucleation in real time using purified proteins. We report that BRCA2 nucleates and stabilizes RAD51 on ssDNA either directly or through an unappreciated diffusion-assisted delivery mechanism involving binding to and sliding along dsDNA, which requires the cooperative action of multiple dsDNA-binding modules in BRCA2. Collectively, our work reveals two distinct mechanisms of BRCA2-dependent RAD51 loading onto ssDNA, which we propose are critical for its diverse functions in maintaining genome stability and cancer suppression.
Keywords: BRCA2; DNA damage and repair; Rad51 filament formation; homologous recombination; single-molecule analysis; tumor suppressor protein.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests S.J.B. is a co-founder, VP Science Strategy, and a shareholder of Artios Pharma Ltd. S.J.B. is also a member of the advisory board of Molecular Cell.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

