Tight control of the APP-Mint1 interaction in regulating amyloid production
- PMID: 37499733
- PMCID: PMC10529462
- DOI: 10.1016/j.brainres.2023.148496
Tight control of the APP-Mint1 interaction in regulating amyloid production
Abstract
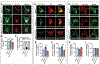
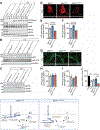
Generation of amyloid-β (Aβ) peptides through the proteolytic processing of the amyloid precursor protein (APP) is a pathogenic event in Alzheimer's disease (AD). APP is a transmembrane protein and endocytosis of APP mediated by the YENPTY motif is a key step in Aβ generation. Mints, a family of cytosolic adaptor proteins, directly bind to the YENPTY motif of APP and facilitate APP trafficking and processing. Here, we generated and examined two Mint1 mutants, Tyr633Ala of Mint1 (Mint1Y633A) that enhanced APP binding, and Tyr549Ala and Phe610Ala mutant (Mint1Y549A/F610A), that reduced APP binding. We investigated how perturbing the APP-Mint1 interaction through these Mint1 mutants alter APP and Mint1 cellular dynamics and Mint1's interaction with its other binding partners. We found that Mint1Y633A increased binding affinity specifically for APP and presenilin1 (catalytic subunit of γ-secretase), that subsequently enhanced APP endocytosis in primary murine neurons. Conversely, Mint1Y549A/F610A exhibited reduced APP affinity and Aβ secretion. The effect of Mint1Y549A/F610A on Aβ release was greater compared to knocking down all three Mint proteins supporting the APP-Mint1 interaction is a critical factor in Aβ production. Altogether, this study highlights the potential of targeting the APP-Mint1 interaction as a therapeutic strategy for AD.
Keywords: APP; Alzheimer’s disease; Amyloid; Mint1.
Copyright © 2023 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

References
-
- Bartling CRO, Jensen TMT, Henry SM, Colliander AL, Sereikaite V, Wenzler M, Jain P, Maric HM, Harpsoe K, Pedersen SW, Clemmensen LS, Haugaard-Kedstrom LM, Gloriam DE, Ho A, Stromgaard K, 2021. Targeting the APP-Mint2 Protein-Protein Interaction with a Peptide-Based Inhibitor Reduces Amyloid-beta Formation. J Am Chem Soc. 143, 891–901. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

