Hsa_circ_0001402 alleviates vascular neointimal hyperplasia through a miR-183-5p-dependent regulation of vascular smooth muscle cell proliferation, migration, and autophagy
- PMID: 37499939
- PMCID: PMC11156604
- DOI: 10.1016/j.jare.2023.07.010
Hsa_circ_0001402 alleviates vascular neointimal hyperplasia through a miR-183-5p-dependent regulation of vascular smooth muscle cell proliferation, migration, and autophagy
Erratum in
-
Corrigendum to "Hsa_circ_0001402 alleviates vascular neointimal hyperplasia through a miR-183-5p-dependent regulation of vascular smooth muscle cell proliferation, migration, and autophagy" [J. Adv. Res. 60 (2024) 93-110].J Adv Res. 2025 Jul;73:761-763. doi: 10.1016/j.jare.2025.03.048. Epub 2025 Mar 25. J Adv Res. 2025. PMID: 40147626 Free PMC article. No abstract available.
Abstract
Introduction: Vascular neointimal hyperplasia, a pathological process observed in cardiovascular diseases such as atherosclerosis and pulmonary hypertension, involves the abundant presence of vascular smooth muscle cells (VSMCs). The proliferation, migration, and autophagy of VSMCs are associated with the development of neointimal lesions. Circular RNAs (circRNAs) play critical roles in regulating VSMC proliferation and migration, thereby participating in neointimal hyperplasia. However, the regulatory roles of circRNAs in VSMC autophagy remain unclear.
Objectives: We aimed to identify circRNAs that are involved in VSMC autophagy-mediated neointimal hyperplasia, as well as elucidate the underlying mechanisms.
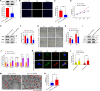
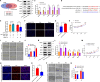
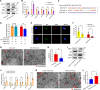
Methods: Dual-luciferase reporter gene assay was performed to validate two competing endogenous RNA axes, hsa_circ_0001402/miR-183-5p/FKBP prolyl isomerase like (FKBPL) and hsa_circ_0001402/miR-183-5p/beclin 1 (BECN1). Cell proliferation and migration analyses were employed to investigate the effects of hsa_circ_0001402, miR-183-5p, or FKBPL on VSMC proliferation and migration. Cell autophagy analysis was conducted to reveal the role of hsa_circ_0001402 or miR-183-5p on VSMC autophagy. The role of hsa_circ_0001402 or miR-183-5p on neointimal hyperplasia was evaluated using a mouse model of common carotid artery ligation.
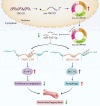
Results: Hsa_circ_0001402 acted as a sponge for miR-183-5p, leading to the suppression of miR-183-5p expression. Through direct interaction with the coding sequence (CDS) of FKBPL, miR-183-5p promoted VSMC proliferation and migration by decreasing FKBPL levels. Besides, miR-183-5p reduced BECN1 levels by targeting the 3'-untranslated region (UTR) of BECN1, thus inhibiting VSMC autophagy. By acting as a miR-183-5p sponge, overexpression of hsa_circ_0001402 increased FKBPL levels to inhibit VSMC proliferation and migration, while simultaneously elevating BECN1 levels to activate VSMC autophagy, thereby alleviating neointimal hyperplasia.
Conclusion: Hsa_circ_0001402, acting as a miR-183-5p sponge, increases FKBPL levels to inhibit VSMC proliferation and migration, while enhancing BECN1 levels to activate VSMC autophagy, thus alleviating neointimal hyperplasia. The hsa_circ_0001402/miR-183-5p/FKBPL axis and hsa_circ_0001402/miR-183-5p/BECN1 axis may offer potential therapeutic targets for neointimal hyperplasia.
Keywords: Autophagy; Migration; Neointima hyperplasia; Proliferation; Vascular smooth muscle cell; hsa_circ_0001402; miR-183-5p.
Copyright © 2024. Production and hosting by Elsevier B.V.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

