Plasma proteome of growing tumors
- PMID: 37500700
- PMCID: PMC10374562
- DOI: 10.1038/s41598-023-38079-9
Plasma proteome of growing tumors
Abstract
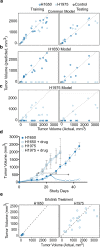
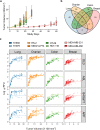
Early detection of cancer is vital for the best chance of successful treatment, but half of all cancers are diagnosed at an advanced stage. A simple and reliable blood screening test applied routinely would therefore address a major unmet medical need. To gain insight into the value of protein biomarkers in early detection and stratification of cancer we determined the time course of changes in the plasma proteome of mice carrying transplanted human lung, breast, colon, or ovarian tumors. For protein measurements we used an aptamer-based assay which simultaneously measures ~ 5000 proteins. Along with tumor lineage-specific biomarkers, we also found 15 markers shared among all cancer types that included the energy metabolism enzymes glyceraldehyde-3-phosphate dehydrogenase, glucose-6-phophate isomerase and dihydrolipoyl dehydrogenase as well as several important biomarkers for maintaining protein, lipid, nucleotide, or carbohydrate balance such as tryptophanyl t-RNA synthetase and nucleoside diphosphate kinase. Using significantly altered proteins in the tumor bearing mice, we developed models to stratify tumor types and to estimate the minimum detectable tumor volume. Finally, we identified significantly enriched common and unique biological pathways among the eight tumor cell lines tested.
© 2023. The Author(s).
Conflict of interest statement
S.G., M.J.W., D.G.A., S.J.W., D.J.S., D.W.D., and N.J. are employees and/or stakeholders of SomaLogic. P.W., C.M., D.C.W. are paid consultants of SomaLogic. A.K.-F. and L.E.H. declare no competing interests.
Figures






References
-
- Alix-Panabières C, Pantel K. Liquid biopsy: From discovery to clinical application. Cancer Discov. 2021;11:858–873. doi: 10.1158/2159-8290.Cd-20-1311. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

