Landscape of mSWI/SNF chromatin remodeling complex perturbations in neurodevelopmental disorders
- PMID: 37500730
- PMCID: PMC10412456
- DOI: 10.1038/s41588-023-01451-6
Landscape of mSWI/SNF chromatin remodeling complex perturbations in neurodevelopmental disorders
Abstract
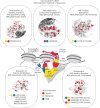
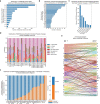
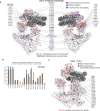
DNA sequencing-based studies of neurodevelopmental disorders (NDDs) have identified a wide range of genetic determinants. However, a comprehensive analysis of these data, in aggregate, has not to date been performed. Here, we find that genes encoding the mammalian SWI/SNF (mSWI/SNF or BAF) family of ATP-dependent chromatin remodeling protein complexes harbor the greatest number of de novo missense and protein-truncating variants among nuclear protein complexes. Non-truncating NDD-associated protein variants predominantly disrupt the cBAF subcomplex and cluster in four key structural regions associated with high disease severity, including mSWI/SNF-nucleosome interfaces, the ATPase-core ARID-armadillo repeat (ARM) module insertion site, the Arp module and DNA-binding domains. Although over 70% of the residues perturbed in NDDs overlap with those mutated in cancer, ~60% of amino acid changes are NDD-specific. These findings provide a foundation to functionally group variants and link complex aberrancies to phenotypic severity, serving as a resource for the chromatin, clinical genetics and neurodevelopment communities.
© 2023. The Author(s).
Conflict of interest statement
C.K. is the scientific founder, scientific advisor to the Board of Directors, scientific advisory board member, shareholder and consultant for Foghorn Therapeutics. C.K. is also a member of the scientific advisory board and is a shareholder of Nested Therapeutics, Nereid Therapeutics and Accent Therapeutics, serves on the scientific advisory board for Fibrogen and serves as a consultant for Google Ventures and Cell Signaling Technologies. C.K. and A.M.V. hold patents in the field of mSWI/SNF complex targeting therapeutics. S.A.S.V. is a member of the scientific advisory board at Ambry Genetics, for which no compensation is received. The other authors declare no competing interests.
Figures








References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

