Enzyme-less nanopore detection of post-translational modifications within long polypeptides
- PMID: 37500774
- PMCID: PMC10656283
- DOI: 10.1038/s41565-023-01462-8
Enzyme-less nanopore detection of post-translational modifications within long polypeptides
Abstract
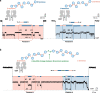
Means to analyse cellular proteins and their millions of variants at the single-molecule level would uncover substantial information previously unknown to biology. Nanopore technology, which underpins long-read DNA and RNA sequencing, holds potential for full-length proteoform identification. We use electro-osmosis in an engineered charge-selective nanopore for the non-enzymatic capture, unfolding and translocation of individual polypeptides of more than 1,200 residues. Unlabelled thioredoxin polyproteins undergo transport through the nanopore, with directional co-translocational unfolding occurring unit by unit from either the C or N terminus. Chaotropic reagents at non-denaturing concentrations accelerate the analysis. By monitoring the ionic current flowing through the nanopore, we locate post-translational modifications deep within the polypeptide chains, laying the groundwork for compiling inventories of the proteoforms in cells and tissues.
© 2023. The Author(s).
Conflict of interest statement
H.B. is the founder of, a consultant for and a shareholder of Oxford Nanopore Technologies, a company engaged in the development of nanopore sensing and sequencing technologies. P.M.-B., W.-H.L., Y.Q., M.R.-R., H.B. and S.G.-M. have filed patents describing the electro-osmotically active nanopores and their applications in proteoform characterization. S.B. was listed as a contributor to the intellectual property.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

