Enhancing pDNA Delivery with Hydroquinine Polymers by Modulating Structure and Composition
- PMID: 37502160
- PMCID: PMC10369409
- DOI: 10.1021/jacsau.3c00126
Enhancing pDNA Delivery with Hydroquinine Polymers by Modulating Structure and Composition
Abstract
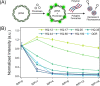
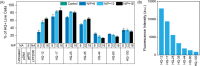
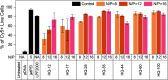
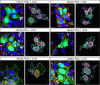
Quinine is a promising natural product building block for polymer-based nucleic acid delivery vehicles as its structure enables DNA binding through both intercalation and electrostatic interactions. However, studies exploring the potential of quinine-based polymers for nucleic acid delivery applications (transfection) are limited. In this work, we used a hydroquinine-functionalized monomer, HQ, with 2-hydroxyethyl acrylate to create a family of seven polymers (HQ-X, X = mole percentage of HQ), with mole percentages of HQ ranging from 12 to 100%. We developed a flow cytometer-based assay for studying the polymer-pDNA complexes (polyplex particles) directly and demonstrate that polymer composition and monomer structure influence polyplex characteristics such as the pDNA loading and the extent of adsorption of serum proteins on polyplex particles. Biological delivery experiments revealed that maximum transgene expression, outperforming commercial controls, was achieved with HQ-25 and HQ-35 as these two variants sustained gene expression over 96 h. HQ-44, HQ-60, and HQ-100 were not successful in inducing transgene expression, despite being able to deliver pDNA into the cells, highlighting that the release of pDNA is likely the bottleneck in transfection for polymers with higher HQ content. Using confocal imaging, we quantified the extent of colocalization between pDNA and lysosomes, proving the remarkable endosomal escape capabilities of the HQ-X polymers. Overall, this study demonstrates the advantages of HQ-X polymers as well as provides guiding principles for improving the monomer structure and polymer composition, supporting the development of the next generation of polymer-based nucleic acid delivery vehicles harnessing the power of natural products.
© 2023 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



Similar articles
-
Binary Copolymer Blending Enhances pDNA Delivery Performance and Colloidal Shelf Stability of Quinine-Based Polyplexes.Bioconjug Chem. 2025 Apr 16;36(4):770-781. doi: 10.1021/acs.bioconjchem.5c00040. Epub 2025 Mar 11. Bioconjug Chem. 2025. PMID: 40067683
-
Quinine-Based Polymers Are Versatile and Effective Vehicles for Intracellular pDNA, mRNA, and Cas9 Protein Delivery.Biomacromolecules. 2024 Oct 14;25(10):6693-6707. doi: 10.1021/acs.biomac.4c00925. Epub 2024 Sep 26. Biomacromolecules. 2024. PMID: 39324490
-
Quinine copolymer reporters promote efficient intracellular DNA delivery and illuminate a protein-induced unpackaging mechanism.Proc Natl Acad Sci U S A. 2020 Dec 29;117(52):32919-32928. doi: 10.1073/pnas.2016860117. Epub 2020 Dec 14. Proc Natl Acad Sci U S A. 2020. PMID: 33318196 Free PMC article.
-
Polymers for nucleic acid transfer-an overview.Adv Genet. 2014;88:231-61. doi: 10.1016/B978-0-12-800148-6.00008-0. Adv Genet. 2014. PMID: 25409608 Review.
-
Progress and prospects of polyplex nanomicelles for plasmid DNA delivery.Curr Gene Ther. 2011 Dec;11(6):457-65. doi: 10.2174/156652311798192879. Curr Gene Ther. 2011. PMID: 22023475 Review.
Cited by
-
Covalent recruitment of polymers and nanoparticles onto glycan-engineered cells enhances gene delivery during short exposure.Chem Sci. 2024 Sep 9;15(38):15731-6. doi: 10.1039/d4sc03666b. Online ahead of print. Chem Sci. 2024. PMID: 39263661 Free PMC article.
-
Cinchona alkaloid copolymers as fluorimetric INHIBIT and colorimetric AND logic gates for detection of iodide.RSC Adv. 2025 Apr 8;15(14):11121-11127. doi: 10.1039/d5ra01281c. eCollection 2025 Apr 4. RSC Adv. 2025. PMID: 40201211 Free PMC article.
References
-
- American Society of Gene & Cell Therapy . Gene, Cell, and RNA Therapy Landscape. Q4 2022 Quarterly Data Report, https://asgct.org/global/documents/asgct_citeline-q4-2022-report_final.aspx (accessed 27 Feb, 2023).
LinkOut - more resources
Full Text Sources
