Preclinical evaluation of PHH-1V vaccine candidate against SARS-CoV-2 in non-human primates
- PMID: 37502366
- PMCID: PMC10299950
- DOI: 10.1016/j.isci.2023.107224
Preclinical evaluation of PHH-1V vaccine candidate against SARS-CoV-2 in non-human primates
Abstract
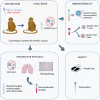
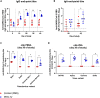
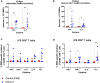
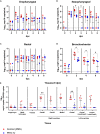
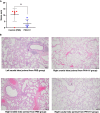
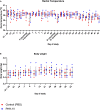
SARS-CoV-2 emerged in December 2019 and quickly spread worldwide, continuously striking with an unpredictable evolution. Despite the success in vaccine production and mass vaccination programs, the situation is not still completely controlled, and therefore accessible second-generation vaccines are required to mitigate the pandemic. We previously developed an adjuvanted vaccine candidate coded PHH-1V, based on a heterodimer fusion protein comprising the RBD domain of two SARS-CoV-2 variants. Here, we report data on the efficacy, safety, and immunogenicity of PHH-1V in cynomolgus macaques. PHH-1V prime-boost vaccination induces high levels of RBD-specific IgG binding and neutralizing antibodies against several SARS-CoV-2 variants, as well as a balanced Th1/Th2 cellular immune response. Remarkably, PHH-1V vaccination prevents SARS-CoV-2 replication in the lower respiratory tract and significantly reduces viral load in the upper respiratory tract after an experimental infection. These results highlight the potential use of the PHH-1V vaccine in humans, currently undergoing Phase III clinical trials.
Keywords: Health sciences; Immune response; Immunology; Microbiology.
© 2023 The Authors.
Conflict of interest statement
Authors indicated as “1” are employees of HIPRA, a private pharmaceutical company that develops and manufactures biological medicines such as vaccines. IrsiCaixa, UPF, Cynbiose, CIRI, VirNext, and CReSA have received financial support from HIPRA. Several patent applications have been filed by HIPRA Scientific S.L.U. and Laboratorios HIPRA, S.A. on different SARS-CoV-2 vaccine candidates and SARS-CoV-2 subunit vaccines, including the novel recombinant RBD fusion heterodimer PHH-1V. A.B., A.P., L.G., L.F., E.P., J.P., T.P., and C.G. are the inventors of these patent applications.
Figures


References
-
- WHO Weekly epidemiological update on COVID-19-14 September 2022. 2022. https://www.who.int/publications/m/item/weekly-epidemiological-update-on...
-
- Goepfert P.A., Fu B., Chabanon A.L., Bonaparte M.I., Davis M.G., Essink B.J., Frank I., Haney O., Janosczyk H., Keefer M.C., et al. Safety and immunogenicity of SARS-CoV-2 recombinant protein vaccine formulations in healthy adults: interim results of a randomised, placebo-controlled, phase 1-2, dose-ranging study. Lancet Infect. Dis. 2021;21:1257–1270. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

