Single-cell reference mapping to construct and extend cell-type hierarchies
- PMID: 37502708
- PMCID: PMC10370450
- DOI: 10.1093/nargab/lqad070
Single-cell reference mapping to construct and extend cell-type hierarchies
Abstract
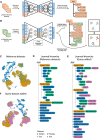
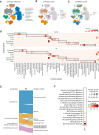
Single-cell genomics is now producing an ever-increasing amount of datasets that, when integrated, could provide large-scale reference atlases of tissue in health and disease. Such large-scale atlases increase the scale and generalizability of analyses and enable combining knowledge generated by individual studies. Specifically, individual studies often differ regarding cell annotation terminology and depth, with different groups specializing in different cell type compartments, often using distinct terminology. Understanding how these distinct sets of annotations are related and complement each other would mark a major step towards a consensus-based cell-type annotation reflecting the latest knowledge in the field. Whereas recent computational techniques, referred to as 'reference mapping' methods, facilitate the usage and expansion of existing reference atlases by mapping new datasets (i.e. queries) onto an atlas; a systematic approach towards harmonizing dataset-specific cell-type terminology and annotation depth is still lacking. Here, we present 'treeArches', a framework to automatically build and extend reference atlases while enriching them with an updatable hierarchy of cell-type annotations across different datasets. We demonstrate various use cases for treeArches, from automatically resolving relations between reference and query cell types to identifying unseen cell types absent in the reference, such as disease-associated cell states. We envision treeArches enabling data-driven construction of consensus atlas-level cell-type hierarchies and facilitating efficient usage of reference atlases.
© The Author(s) 2023. Published by Oxford University Press on behalf of NAR Genomics and Bioinformatics.
Figures


Similar articles
-
Efficient and precise single-cell reference atlas mapping with Symphony.Nat Commun. 2021 Oct 7;12(1):5890. doi: 10.1038/s41467-021-25957-x. Nat Commun. 2021. PMID: 34620862 Free PMC article.
-
Mapping single-cell data to reference atlases by transfer learning.Nat Biotechnol. 2022 Jan;40(1):121-130. doi: 10.1038/s41587-021-01001-7. Epub 2021 Aug 30. Nat Biotechnol. 2022. PMID: 34462589 Free PMC article.
-
scATAnno: Automated Cell Type Annotation for single-cell ATAC Sequencing Data.bioRxiv [Preprint]. 2024 Mar 25:2023.06.01.543296. doi: 10.1101/2023.06.01.543296. bioRxiv. 2024. PMID: 37333088 Free PMC article. Preprint.
-
Considerations for building and using integrated single-cell atlases.Nat Methods. 2025 Jan;22(1):41-57. doi: 10.1038/s41592-024-02532-y. Epub 2024 Dec 13. Nat Methods. 2025. PMID: 39672979 Review.
-
Single-Cell Analysis for Whole-Organism Datasets.Annu Rev Biomed Data Sci. 2021 Jul 20;4:207-226. doi: 10.1146/annurev-biodatasci-092820-031008. Epub 2021 May 11. Annu Rev Biomed Data Sci. 2021. PMID: 34465173 Review.
Cited by
-
Benchmarking cross-species single-cell RNA-seq data integration methods: towards a cell type tree of life.Nucleic Acids Res. 2025 Jan 7;53(1):gkae1316. doi: 10.1093/nar/gkae1316. Nucleic Acids Res. 2025. PMID: 39778870 Free PMC article.
-
scPML: pathway-based multi-view learning for cell type annotation from single-cell RNA-seq data.Commun Biol. 2023 Dec 14;6(1):1268. doi: 10.1038/s42003-023-05634-z. Commun Biol. 2023. PMID: 38097699 Free PMC article.
-
BrainCellR: A precise cell type nomenclature pipeline for comparative analysis across brain single-cell datasets.Comput Struct Biotechnol J. 2024 Nov 26;23:4306-4314. doi: 10.1016/j.csbj.2024.11.038. eCollection 2024 Dec. Comput Struct Biotechnol J. 2024. PMID: 39687760 Free PMC article.
-
CellWalker2: multi-omic discovery of hierarchical cell type relationships and their associations with genomic annotations.bioRxiv [Preprint]. 2024 May 17:2024.05.17.594770. doi: 10.1101/2024.05.17.594770. bioRxiv. 2024. Update in: Cell Genom. 2025 Jul 9;5(7):100886. doi: 10.1016/j.xgen.2025.100886. PMID: 38798605 Free PMC article. Updated. Preprint.
-
Profiling cell identity and tissue architecture with single-cell and spatial transcriptomics.Nat Rev Mol Cell Biol. 2025 Jan;26(1):11-31. doi: 10.1038/s41580-024-00768-2. Epub 2024 Aug 21. Nat Rev Mol Cell Biol. 2025. PMID: 39169166 Review.
References
Associated data
LinkOut - more resources
Full Text Sources

