This is a preprint.
Strong Positive Selection Biases Identity-By-Descent-Based Inferences of Recent Demography and Population Structure in Plasmodium falciparum
- PMID: 37502843
- PMCID: PMC10370022
- DOI: 10.1101/2023.07.14.549114
Strong Positive Selection Biases Identity-By-Descent-Based Inferences of Recent Demography and Population Structure in Plasmodium falciparum
Update in
-
Strong positive selection biases identity-by-descent-based inferences of recent demography and population structure in Plasmodium falciparum.Nat Commun. 2024 Mar 20;15(1):2499. doi: 10.1038/s41467-024-46659-0. Nat Commun. 2024. PMID: 38509066 Free PMC article.
Abstract
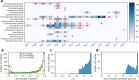
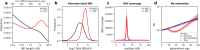
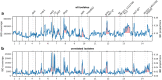
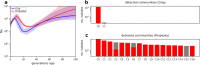
Malaria genomic surveillance often estimates parasite genetic relatedness using metrics such as Identity-By-Decent (IBD). Yet, strong positive selection stemming from antimalarial drug resistance or other interventions may bias IBD-based estimates. In this study, we utilized simulations, a true IBD inference algorithm, and empirical datasets from different malaria transmission settings to investigate the extent of such bias and explore potential correction strategies. We analyzed whole genome sequence data generated from 640 new and 4,026 publicly available Plasmodium falciparum clinical isolates. Our findings demonstrated that positive selection distorts IBD distributions, leading to underestimated effective population size and blurred population structure. Additionally, we discovered that the removal of IBD peak regions partially restored the accuracy of IBD-based inferences, with this effect contingent on the population's background genetic relatedness. Consequently, we advocate for selection correction for parasite populations undergoing strong, recent positive selection, particularly in high malaria transmission settings.
Keywords: Effective population size; Genetic relatedness; Identity-By-Descent; Malaria; Population structure; Positive selection.
Figures


References
-
- World Health Organization. World malaria report 2022. (World Health Organization, 2022).
-
- Packard R. M. The origins of antimalarial-drug resistance. N. Engl. J. Med. 371, 397–9 (2014). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
