This is a preprint.
De novo purine metabolism is a metabolic vulnerability of cancers with low p16 expression
- PMID: 37503050
- PMCID: PMC10369956
- DOI: 10.1101/2023.07.15.549149
De novo purine metabolism is a metabolic vulnerability of cancers with low p16 expression
Update in
-
De Novo Purine Metabolism is a Metabolic Vulnerability of Cancers with Low p16 Expression.Cancer Res Commun. 2024 May 2;4(5):1174-1188. doi: 10.1158/2767-9764.CRC-23-0450. Cancer Res Commun. 2024. PMID: 38626341 Free PMC article.
Abstract
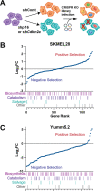
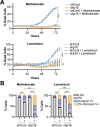
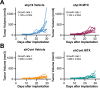
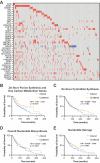
p16 is a tumor suppressor encoded by the CDKN2A gene whose expression is lost in ~50% of all human cancers. In its canonical role, p16 inhibits the G1-S phase cell cycle progression through suppression of cyclin dependent kinases. Interestingly, p16 also has roles in metabolic reprogramming, and we previously published that loss of p16 promotes nucleotide synthesis via the pentose phosphate pathway. Whether other nucleotide metabolic genes and pathways are affected by p16/CDKN2A loss and if these can be specifically targeted in p16/CDKN2A-low tumors has not been previously explored. Using CRISPR KO libraries in multiple isogenic human and mouse melanoma cell lines, we determined that many nucleotide metabolism genes are negatively enriched in p16/CDKN2A knockdown cells compared to controls. Indeed, many of the genes that are required for survival in the context of low p16/CDKN2A expression based on our CRISPR screens are upregulated in p16 knockdown melanoma cells and those with endogenously low CDKN2A expression. We determined that cells with low p16/Cdkn2a expression are sensitive to multiple inhibitors of de novo purine synthesis, including anti-folates. Tumors with p16 knockdown were more sensitive to the anti-folate methotrexate in vivo than control tumors. Together, our data provide evidence to reevaluate the utility of these drugs in patients with p16/CDKN2A-low tumors as loss of p16/CDKN2A may provide a therapeutic window for these agents.
Keywords: anti-folates; anti-metabolites; folate metabolism; melanoma; one carbon metabolism.
Conflict of interest statement
Declaration of Interests In the past three years, C.A.L. has consulted for Astellas Pharmaceuticals, Odyssey Therapeutics, Third Rock Ventures, and T-Knife Therapeutics, and is an inventor on patents pertaining to Kras regulated metabolic pathways, redox control pathways in pancreatic cancer, and targeting the GOT1-ME1 pathway as a therapeutic approach (US Patent No: 2015126580-A1, 05/07/2015; US Patent No: 20190136238, 05/09/2019; International Patent No: WO2013177426-A2, 04/23/2015). All other authors declare no competing interests.
Figures


References
-
- Abbott K.L., Ali A., Casalena D., Do B.T., Ferreira R., Cheah J.H., Soule C.K., Deik A., Kunchok T., Schmidt D.R., Renner S., Honeder S.E., Wu M., Chan S.H., Tseyang T., Stoltzfus A.T., Michel S.L.J., Greaves D., Hsu P.P., Ng C.W., Zhang C.J., Farsidjani A., Kent J.R., Madariaga M.L.L., Gramatikov I.M.T., Matheson N.J., Lewis C.A., Clish C.B., Rees M.G., Roth J.A., Griner L.M., Muir A., Auld D.S., and Vander Heiden M.G.. 2023. Screening in serum-derived medium reveals differential response to compounds targeting metabolism. Cell chemical biology. - PMC - PubMed
-
- Alhalabi O., Chen J., Zhang Y., Lu Y., Wang Q., Ramachandran S., Tidwell R.S., Han G., Yan X., Meng J., Wang R., Hoang A.G., Wang W.L., Song J., Lopez L., Andreev-Drakhlin A., Siefker-Radtke A., Zhang X., Benedict W.F., Shah A.Y., Wang J., Msaouel P., Zhang M., Guo C.C., Czerniak B., Behrens C., Soto L., Papadimitrakopoulou V., Lewis J., Rinsurongkawong W., Rinsurongkawong V., Lee J., Roth J., Swisher S., Wistuba I., Heymach J., Wang J., Campbell M.T., Efstathiou E., Titus M., Logothetis C.J., Ho T.H., Zhang J., Wang L., and Gao J.. 2022. MTAP deficiency creates an exploitable target for antifolate therapy in 9p21-loss cancers. Nature communications. 13:1797. - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
