This is a preprint.
Phase separation of ecDNA aggregates establishes in-trans contact domains boosting selective MYC regulatory interactions
- PMID: 37503084
- PMCID: PMC10370113
- DOI: 10.1101/2023.07.17.549291
Phase separation of ecDNA aggregates establishes in-trans contact domains boosting selective MYC regulatory interactions
Abstract
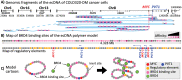
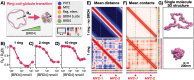
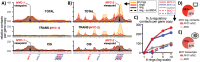
Extrachromosomal DNAs (ecDNAs) are found in the nucleus of an array of human cancer cells where they can form clusters that were associated to oncogene overexpression, as they carry genes and cis-regulatory elements. Yet, the mechanisms of aggregation and gene amplification beyond copy-number effects remain mostly unclear. Here, we investigate, at the single molecule level, MYC-harboring ecDNAs of COLO320-DM colorectal cancer cells by use of a minimal polymer model of the interactions of ecDNA BRD4 binding sites and BRD4 molecules. We find that BRD4 induces ecDNAs phase separation, resulting in the self-assembly of clusters whose predicted structure is validated against HiChIP data (Hung et al., 2021). Clusters establish in-trans associated contact domains (I-TADs) enriched, beyond copy number, in regulatory contacts among specific ecDNA regions, encompassing its PVT1-MYC fusions but not its other canonical MYC copy. That explains why the fusions originate most of ecDNA MYC transcripts (Hung et al., 2021), and shows that ecDNA clustering per se is important but not sufficient to amplify oncogene expression beyond copy-number, reconciling opposite views on the role of clusters (Hung et al., 2021; Zhu et al., 2021; Purshouse et al. 2022). Regulatory contacts become strongly enriched as soon as half a dozen ecDNAs aggregate, then saturate because of steric hindrance, highlighting that even cells with few ecDNAs can experience pathogenic MYC upregulations. To help drug design and therapeutic applications, with the model we dissect the effects of JQ1, a BET inhibitor. We find that JQ1 reverses ecDNA phase separation hence abolishing I-TADs and extra regulatory contacts, explaining how in COLO320-DM cells it reduces MYC transcription (Hung et al., 2021).
Figures



References
-
- Barbieri M, Xie SQ, Torlai Triglia E, Chiariello AM, Bianco S, de Santiago I, Branco MR, Rueda D, Nicodemi M, Pombo A. Active and poised promoter states drive folding of the extended HoxB locus in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 24, 515–524 (2017). - PubMed
-
- Bianco S, Lupianez DG, Chiariello AM, Annunziatella C, Kraft K, Schoepflin R, Wittler L, Andrey G, Vingron M, Pombo A, Mundlos S, Nicodemi M. Polymer Physics Predicts the Effects of Structural Variants on Chromatin Architecture. Nature Genetics 50, 662 (2018) - PubMed
-
- Brangwynne C., Tompa P. & Pappu R. Polymer physics of intracellular phase transitions. Nature Phys 11, 899–904 (2015). 10.1038/nphys3532 - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
