This is a preprint.
Entire Expressed Peripheral Blood Transcriptome in Pediatric Severe Malarial Anemia
- PMID: 37503086
- PMCID: PMC10371159
- DOI: 10.21203/rs.3.rs-3150748/v1
Entire Expressed Peripheral Blood Transcriptome in Pediatric Severe Malarial Anemia
Update in
-
Entire expressed peripheral blood transcriptome in pediatric severe malarial anemia.Nat Commun. 2024 Jun 12;15(1):5037. doi: 10.1038/s41467-024-48259-4. Nat Commun. 2024. PMID: 38866743 Free PMC article.
Abstract
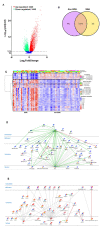
This study on severe malarial anemia (SMA: Hb < 6.0 g/dL), a leading global cause of childhood morbidity and mortality, analyzed the entire expressed transcriptome in whole blood from children with non-SMA (Hb ≥ 6.0 g/dL, n = 41) and SMA (n = 25). Analyses revealed 3,420 up-regulated and 3,442 down-regulated transcripts, signifying impairments in host inflammasome activation, cell death, innate immune responses, and cellular stress responses in SMA. Immune cell profiling showed a decreased antigenic and immune priming response in children with SMA, favoring polarization toward cellular proliferation and repair. Enrichment analysis further identified altered neutrophil and autophagy-related processes, consistent with neutrophil degranulation and altered ubiquitination and proteasome degradation. Pathway analyses highlighted SMA-related alterations in cellular homeostasis, signaling, response to environmental cues, and cellular and immune stress responses. Validation with a qRT-PCR array showed strong concordance with the sequencing data. These findings identify key molecular themes in SMA pathogenesis, providing potential targets for new malaria therapies.
Conflict of interest statement
COMPETING INTERESTS The authors declare no competing interests.
Figures




References
-
- WHO. World malaria report 2022. . (World Health Organization, 2022).
-
- WHO. World malaria report 2021. Report No. 9240040498, (2021).
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

