This is a preprint.
SURROGATE SELECTION OVERSAMPLES EXPANDED T CELL CLONOTYPES
- PMID: 37503118
- PMCID: PMC10369934
- DOI: 10.1101/2023.07.13.548950
SURROGATE SELECTION OVERSAMPLES EXPANDED T CELL CLONOTYPES
Update in
-
SURROGATE SELECTION OVERSAMPLES EXPANDED T CELL CLONOTYPES.Ann Appl Stat. 2025 Sep;19(3):1884-1907. doi: 10.1214/25-aoas2032. Epub 2025 Aug 28. Ann Appl Stat. 2025. PMID: 41036086 Free PMC article.
Abstract
Inference from immunological data on cells in the adaptive immune system may benefit from modeling specifications that describe variation in the sizes of various clonal sub-populations. We develop one such specification in order to quantify the effects of surrogate selection assays, which we confirm may lead to an enrichment for amplified, potentially disease-relevant cell clones. Our specification couples within-clonotype birth-death processes with an exchangeable model across clonotypes. Beyond enrichment questions about the surrogate selection design, our framework enables a study of sampling properties of elementary sample diversity statistics; it also points to new statistics that may usefully measure the burden of somatic genomic alterations associated with clonal expansion. We examine statistical properties of immunological samples governed by the coupled model specification, and we illustrate calculations in surrogate selection studies of melanoma and in single-cell genomic studies of cell repertoires.
Keywords: Bayes’s rule; Yule-Simon law; clonal expansion; diversity statistic; enrichment; exchangeable birth-death processes; experimental design; single cell sequencing; size bias; somatic mutation.
Figures





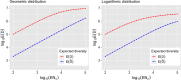

References
-
- Albertini R. J. (2001). HPRT mutations in humans: biomarkers for mechanistic studies. Mutation Research/Reviews in Mutation Research 489 1–16. - PubMed
-
- Albertini R. J., Nicklas J. A., O’Neill J. P. and Robison S. H. (1990). In vivo somatic mutations in humans: measurement and analysis. Annual review of genetics 24 305–326. - PubMed
-
- Aldous D. (1996). Probability Distributions on Cladograms. In Random Discrete Structures 1–18. Springer.
-
- Angerer W. P. (2001). An explicit representation of the Luria-Delbrück distribution. Journal of mathematical biology 42 145–174. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
