This is a preprint.
A modular chemigenetic calcium indicator enables in vivo functional imaging with near-infrared light
- PMID: 37503182
- PMCID: PMC10370049
- DOI: 10.1101/2023.07.18.549527
A modular chemigenetic calcium indicator enables in vivo functional imaging with near-infrared light
Update in
-
A modular chemigenetic calcium indicator for multiplexed in vivo functional imaging.Nat Methods. 2024 Oct;21(10):1916-1925. doi: 10.1038/s41592-024-02411-6. Epub 2024 Sep 20. Nat Methods. 2024. PMID: 39304767 Free PMC article.
Abstract
Genetically encoded fluorescent calcium indicators have revolutionized neuroscience and other biological fields by allowing cellular-resolution recording of physiology during behavior. However, we currently lack bright, genetically targetable indicators in the near infrared that can be used in animals. Here, we describe WHaloCaMP, a modular chemigenetic calcium indicator built from bright dye-ligands and protein sensor domains that can be genetically targeted to specific cell populations. Fluorescence change in WHaloCaMP results from reversible quenching of the bound dye via a strategically placed tryptophan. WHaloCaMP is compatible with rhodamine dye-ligands that fluoresce from green to near-infrared, including several dye-ligands that efficiently label the central nervous system in animals. When bound to a near-infrared dye-ligand, WHaloCaMP1a is more than twice as bright as jGCaMP8s, and shows a 7× increase in fluorescence intensity and a 2.1 ns increase in fluorescence lifetime upon calcium binding. We use WHaloCaMP1a with near-infrared fluorescence emission to image Ca2+ responses in flies and mice, to perform three-color multiplexed functional imaging of hundreds of neurons and astrocytes in zebrafish larvae, and to quantitate calcium concentration using fluorescence lifetime imaging microscopy (FLIM).
Conflict of interest statement
Competing interests H. F. and E. R. S. have filed patent applications on tryptophan-containing chemigenetic fluorescent indicators. L. D. L. and J. B. G have filed patents and patent applications on fluorinated and azetidine-containing rhodamines.
Figures

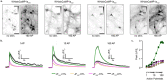


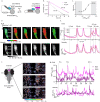
References
-
- O’Banion C.P. & Yasuda R. Fluorescent sensors for neuronal signaling. Current Opinion in Neurobiology 63, 31–41 (2020). - PubMed
-
- Sabatini B.L. & Tian L. Imaging neurotransmitter and neuromodulator dynamics in vivo with genetically encoded indicators. Neuron 108, 17–32 (2020). - PubMed
-
- Liu P., Mu X., Zhang X.-D. & Ming D. The near-infrared-II fluorophores and advanced microscopy technologies development and application in bioimaging. Bioconjugate Chemistry 31, 260–275 (2020). - PubMed
-
- Farrants H. & Tebo A.G. Fluorescent chemigenetic actuators and indicators for use in living animals. Current Opinion in Pharmacology 62, 159–167 (2022). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
