This is a preprint.
Gene expression variation underlying tissue-specific responses to copper stress in Drosophila melanogaster
- PMID: 37503205
- PMCID: PMC10370140
- DOI: 10.1101/2023.07.12.548746
Gene expression variation underlying tissue-specific responses to copper stress in Drosophila melanogaster
Update in
-
Gene expression variation underlying tissue-specific responses to copper stress in Drosophila melanogaster.G3 (Bethesda). 2024 Mar 6;14(3):jkae015. doi: 10.1093/g3journal/jkae015. G3 (Bethesda). 2024. PMID: 38262701 Free PMC article.
Abstract
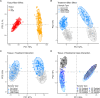
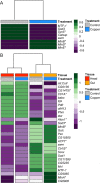
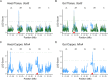
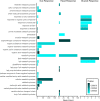
Copper is one of a handful of biologically necessary heavy metals that is also a common environmental pollutant. Under normal conditions, copper ions are required for many key physiological processes. However, in excess, copper quickly results in cell and tissue damage that can range in severity from temporary injury to permanent neurological damage. Because of its biological relevance, and because many conserved copper-responsive genes also respond to other non-essential heavy metal pollutants, copper resistance in Drosophila melanogaster is a useful model system with which to investigate the genetic control of the response to heavy metal stress. Because heavy metal toxicity has the potential to differently impact specific tissues, we genetically characterized the control of the gene expression response to copper stress in a tissue-specific manner in this study. We assessed the copper stress response in head and gut tissue of 96 inbred strains from the Drosophila Synthetic Population Resource (DSPR) using a combination of differential expression analysis and expression quantitative trait locus (eQTL) mapping. Differential expression analysis revealed clear patterns of tissue-specific expression, primarily driven by a more pronounced gene expression response in gut tissue. eQTL mapping of gene expression under control and copper conditions as well as for the change in gene expression following copper exposure (copper response eQTL) revealed hundreds of genes with tissue-specific local cis-eQTL and many distant trans-eQTL. eQTL associated with MtnA, Mdr49, Mdr50, and Sod3 exhibited genotype by environment effects on gene expression under copper stress, illuminating several tissue- and treatment-specific patterns of gene expression control. Together, our data build a nuanced description of the roles and interactions between allelic and expression variation in copper-responsive genes, provide valuable insight into the genomic architecture of susceptibility to metal toxicity, and highlight many candidate genes for future functional characterization.
Keywords: DSPR; eQTL mapping; gene expression; heavy metals.
Conflict of interest statement
Conflict of Interest The authors declare no conflicts of interest.
Figures



Similar articles
-
Gene expression variation underlying tissue-specific responses to copper stress in Drosophila melanogaster.G3 (Bethesda). 2024 Mar 6;14(3):jkae015. doi: 10.1093/g3journal/jkae015. G3 (Bethesda). 2024. PMID: 38262701 Free PMC article.
-
Characterizing the genetic basis of copper toxicity in Drosophila reveals a complex pattern of allelic, regulatory, and behavioral variation.Genetics. 2021 Mar 3;217(1):1-20. doi: 10.1093/genetics/iyaa020. Genetics. 2021. PMID: 33683361 Free PMC article.
-
Lead Modulates trans- and cis-Expression Quantitative Trait Loci (eQTLs) in Drosophila melanogaster Heads.Front Genet. 2018 Sep 20;9:395. doi: 10.3389/fgene.2018.00395. eCollection 2018. Front Genet. 2018. PMID: 30294342 Free PMC article.
-
Heavy-metal-induced reactive oxygen species: phytotoxicity and physicochemical changes in plants.Rev Environ Contam Toxicol. 2014;232:1-44. doi: 10.1007/978-3-319-06746-9_1. Rev Environ Contam Toxicol. 2014. PMID: 24984833 Review.
-
Biomarkers in aquatic plants: selection and utility.Rev Environ Contam Toxicol. 2009;198:49-109. doi: 10.1007/978-0-387-09647-6_2. Rev Environ Contam Toxicol. 2009. PMID: 19253039 Review.
References
-
- Genet. 109: 223–239. 10.1016/j.ajhg.2022.01.002 - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
