This is a preprint.
Petascale Homology Search for Structure Prediction
- PMID: 37503235
- PMCID: PMC10369885
- DOI: 10.1101/2023.07.10.548308
Petascale Homology Search for Structure Prediction
Update in
-
Petabase-Scale Homology Search for Structure Prediction.Cold Spring Harb Perspect Biol. 2024 May 2;16(5):a041465. doi: 10.1101/cshperspect.a041465. Cold Spring Harb Perspect Biol. 2024. PMID: 38316555 Review.
Abstract
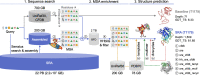
The recent CASP15 competition highlighted the critical role of multiple sequence alignments (MSAs) in protein structure prediction, as demonstrated by the success of the top AlphaFold2-based prediction methods. To push the boundaries of MSA utilization, we conducted a petabase-scale search of the Sequence Read Archive (SRA), resulting in gigabytes of aligned homologs for CASP15 targets. These were merged with default MSAs produced by ColabFold-search and provided to ColabFold-predict. By using SRA data, we achieved highly accurate predictions (GDT_TS > 70) for 66% of the non-easy targets, whereas using ColabFold-search default MSAs scored highly in only 52%. Next, we tested the effect of deep homology search and ColabFold's advanced features, such as more recycles, on prediction accuracy. While SRA homologs were most significant for improving ColabFold's CASP15 ranking from 11th to 3rd place, other strategies contributed too. We analyze these in the context of existing strategies to improve prediction.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
