This is a preprint.
Phenotypic Correlates of Structural and Functional Protein Impairments Resultant from ALDH5A1 Variants
- PMID: 37503297
- PMCID: PMC10371128
- DOI: 10.21203/rs.3.rs-3111263/v1
Phenotypic Correlates of Structural and Functional Protein Impairments Resultant from ALDH5A1 Variants
Update in
-
Phenotypic correlates of structural and functional protein impairments resultant from ALDH5A1 variants.Hum Genet. 2023 Dec;142(12):1755-1776. doi: 10.1007/s00439-023-02613-6. Epub 2023 Nov 14. Hum Genet. 2023. PMID: 37962671
Abstract
Objective: To investigate the genotype-to-protein-to-phenotype correlations of succinic semialdehyde dehydrogenase deficiency (SSADHD), an inherited metabolic disorder of γ-aminobutyric acid catabolism.
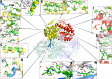
Methods: Bioinformatics and in silico mutagenesis analyses of ALDH5A1 variants were performed to evaluate their impact on protein stability, active site and co-factor binding domains, splicing, and homotetramer formation. Protein abnormalities were then correlated with a validated disease-specific clinical severity score and neurological, neuropsychological, biochemical, neuroimaging, and neurophysiological metrics.
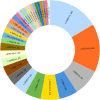
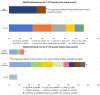
Results: A total of 58 individuals (1:1 male/female ratio) were affected by 32 ALDH5A1 pathogenic variants, eight of which were novel. Compared to individuals with single homotetrameric or multiple homo and heterotetrameric proteins, those predicted not to synthesize any functional enzyme protein had significantly lower expression of ALDH5A1 (p = 0.001), worse overall clinical outcomes (p = 0.008) and specifically more severe cognitive deficits (p = 0.01), epilepsy (p = 0.04) and psychiatric morbidity (p = 0.04). Compared to individuals with predictions of having no protein or a protein impaired in catalytic functions, subjects whose proteins were predicted to be impaired in stability, folding, or oligomerization had a better overall clinical outcome (p = 0.02) and adaptive skills (p = 0.04).
Conclusions: The quantity and type of enzyme proteins (no protein, single homotetramers, or multiple homo and heterotetramers), as well as their structural and functional impairments (catalytic or stability, folding, or oligomerization), contribute to phenotype severity in SSADHD. These findings are valuable for assessment of disease prognosis and management, including patient selection for gene replacement therapy. Furthermore, they provide a roadmap to determine genotype-to-protein-to-phenotype relationships in other autosomal recessive disorders.
Keywords: 4-hydroxybutyricuria; SSADH Deficiency; genotype-phenotype; in-silico; variants.
Conflict of interest statement
Competing Interests The authors I.T.L, J.B.R., M.B., S.C., M.L.D., E.A., T.O., K.J., À.G.C., N.J.P., K.M.G. have no relevant financial or non- financial interests to discolose. The authors A.R. and H.H.C.L. are co-founders and have equity in Galibra Neuroscience, Inc., which develops treatments for SSADH deficiency, including gene replacement therapy mentioned in this study. The authors A.R., H.C.C.L., and P.L.P. are inventors of a filed SSADH deficiency gene therapy patent.
Figures
References
-
- Martin K, McConnell A, Elsea SH. Assessing Prevalence and Carrier Frequency of Succinic Semialdehyde Dehydrogenase Deficiency. J Child Neurol. 2021. Nov;36(13–14):1218–22. - PubMed
-
- Pearl PL, Gibson KM, Acosta MT, et al. Clinical spectrum of succinic semialdehyde dehydrogenase deficiency. Neurology. 2003. May 13;60(9):1413–7. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

