A Prototype Assay Multiplexing SARS-CoV-2 3CL-Protease and Angiotensin-Converting Enzyme 2 for Saliva-Based Diagnostics in COVID-19
- PMID: 37504081
- PMCID: PMC10377347
- DOI: 10.3390/bios13070682
A Prototype Assay Multiplexing SARS-CoV-2 3CL-Protease and Angiotensin-Converting Enzyme 2 for Saliva-Based Diagnostics in COVID-19
Abstract
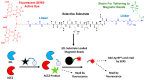
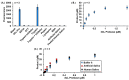
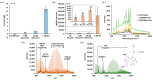
With the current state of COVID-19 changing from a pandemic to being more endemic, the priorities of diagnostics will likely vary from rapid detection to stratification for the treatment of the most vulnerable patients. Such patient stratification can be facilitated using multiple markers, including SARS-CoV-2-specific viral enzymes, like the 3CL protease, and viral-life-cycle-associated host proteins, such as ACE2. To enable future explorations, we have developed a fluorescent and Raman spectroscopic SARS-CoV-2 3CL protease assay that can be run sequentially with a fluorescent ACE2 activity measurement within the same sample. Our prototype assay functions well in saliva, enabling non-invasive sampling. ACE2 and 3CL protease activity can be run with minimal sample volumes in 30 min. To test the prototype, a small initial cohort of eight clinical samples was used to check if the assay could differentiate COVID-19-positive and -negative samples. Though these small clinical cohort samples did not reach statistical significance, results trended as expected. The high sensitivity of the assay also allowed the detection of a low-activity 3CL protease mutant.
Keywords: COVID-19; enzymatic assays; proteases.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- WHO Coronavirus (COVID-19) Dashboard. [(accessed on 16 November 2022)]. Available online: https://covid19.who.int.
-
- Wang H., Paulson K.R., Pease S.A., Watson S., Comfort H., Zheng P., Aravkin A.Y., Bisignano C., Barber R.M., Alam T., et al. Estimating Excess Mortality Due to the COVID-19 Pandemic: A Systematic Analysis of COVID-19-Related Mortality, 2020–2021. Lancet. 2022;399:1513–1536. doi: 10.1016/S0140-6736(21)02796-3. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

