Multiplex PCR-Lateral Flow Dipstick Method for Detection of Thermostable Direct Hemolysin (TDH) Producing V. parahaemolyticus
- PMID: 37504096
- PMCID: PMC10377466
- DOI: 10.3390/bios13070698
Multiplex PCR-Lateral Flow Dipstick Method for Detection of Thermostable Direct Hemolysin (TDH) Producing V. parahaemolyticus
Abstract
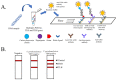
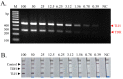
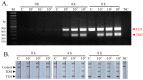
Vibrio parahaemolyticus is usually found in seafood and causes acute gastroenteritis in humans. Therefore, a detection method of pathogenic V. parahaemolyticus is necessary. Multiplex PCR combined with lateral flow dipstick (LFD) assay was developed to detect pathogenic V. parahaemolyticus. Biotin-, FAM-, and Dig-conjugated primers targeting thermolabile hemolysin (TLH) and thermostable direct hemolysin (TDH) genes were used for multiplex PCR amplification. The condition of the method was optimized and evaluated by agarose gel electrophoresis and universal lateral flow dipstick. The specificity assay was evaluated using strains belonging to seven foodborne pathogen species. The sensitivity of the method was also evaluated using DNA in the concentration range of 0.39-100 ng/reaction. The artificial spiking experiment was performed using 10 g of shrimp samples with an enrichment time of 0, 4, and 8 h with 101, 102, and 103 CFU of V. parahaemolyticus. The developed multiplex PCR-LFD assay showed no non-specific amplification with a limit of the detection of 0.78 ng DNA/reaction visualized by agarose gel electrophoresis and 0.39 ng DNA with LFD assay. The artificial spiking experiment demonstrated that this method could detect pathogenic V. parahaemolyticus at 10 CFU/10 g shrimp samples following a 4 h of enrichment. Multiplex PCR-LFD assay was therefore established for detecting pathogenic V. parahaemolyticus with high sensitivity and specificity and might be a useful tool to develop a detection kit used in the food safety sector.
Keywords: PCR; TDH; TLH; V. parahaemolyticus; lateral flow dipstick; seafood.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

