EMT Features in Claudin-Low versus Claudin-Non-Suppressed Breast Cancers and the Role of Epigenetic Modifications
- PMID: 37504297
- PMCID: PMC10378159
- DOI: 10.3390/cimb45070381
EMT Features in Claudin-Low versus Claudin-Non-Suppressed Breast Cancers and the Role of Epigenetic Modifications
Abstract
Background: Breast cancers are heterogeneous and are classified according to the expression of ER, PR and HER2 receptors to distinct groups with prognostic and therapeutic implications. Within the triple-negative group, with no expression of these three receptors, molecular heterogeneity exists but is currently not exploited in the clinic. The claudin-low phenotype is present in a subset of triple-negative breast cancers and constitutes together with basal-like cancers the most extensive groups within triple-negative breast cancers. Suppression of epithelial cell adhesion molecules in claudin-low cancers is also a hallmark of Epithelial Mesenchymal Transition (EMT).
Methods: The groups of claudin-low and claudin-non-suppressed breast cancers from the extensive publicly available genomic cohorts of the METABRIC study were examined to delineate and compare their molecular landscape. Genetic and epigenetic alterations of key factors involved in EMT and potentially associated with the pathogenesis of the claudin-low phenotype were analyzed in the two groups.
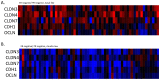
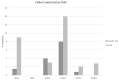
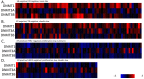
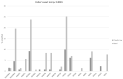
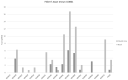
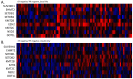
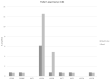
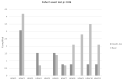
Results: Claudin-low cancers displayed up-regulation of several core transcription factors of EMT at the mRNA level, compared with claudin-non-suppressed breast cancers. Global promoter DNA methylation was increased in both groups of triple-negative cancers and in claudin-low ER-positive cancers compared with the rest of ER-positive cancers. Histone modifier enzymes, including methyltransferases, demethylases, acetyltransferases and deacetylases displayed amplifications more frequently in claudin-non-suppressed triple-negative cancers than in claudin-low counterparts and the expression of some of these enzymes differed significantly between the two groups.
Conclusion: Claudin-low and claudin-non-suppressed triple-negative breast cancers differ in their landscape of EMT core regulators and epigenetic regulators. These differences may be explored as targets for therapeutic interventions specific to the two groups of triple-negative breast cancers.
Keywords: claudins; epigenetic; epithelial-to-mesenchymal transition; methylation; plasticity.
Conflict of interest statement
The author declares no conflict of interest.
Figures




Similar articles
-
Dysregulation of the epigenome in triple-negative breast cancers: basal-like and claudin-low breast cancers express aberrant DNA hypermethylation.Exp Mol Pathol. 2013 Dec;95(3):276-87. doi: 10.1016/j.yexmp.2013.09.001. Epub 2013 Sep 14. Exp Mol Pathol. 2013. PMID: 24045095
-
Comparison of Clinical Subtypes of Breast Cancer within the Claudin-Low Molecular Cluster Reveals Distinct Phenotypes.Cancers (Basel). 2023 May 10;15(10):2689. doi: 10.3390/cancers15102689. Cancers (Basel). 2023. PMID: 37345027 Free PMC article.
-
Molecular Characteristics and Therapeutic Vulnerabilities of Claudin-low Breast Cancers Derived from Cell Line Models.Cancer Genomics Proteomics. 2023 Nov-Dec;20(6):539-555. doi: 10.21873/cgp.20404. Cancer Genomics Proteomics. 2023. PMID: 37889067 Free PMC article.
-
New Insights into the Implication of Epigenetic Alterations in the EMT of Triple Negative Breast Cancer.Cancers (Basel). 2019 Apr 18;11(4):559. doi: 10.3390/cancers11040559. Cancers (Basel). 2019. PMID: 31003528 Free PMC article. Review.
-
Epigenetic Alterations in Triple-Negative Breast Cancer-The Critical Role of Extracellular Matrix.Cancers (Basel). 2021 Feb 9;13(4):713. doi: 10.3390/cancers13040713. Cancers (Basel). 2021. PMID: 33572395 Free PMC article. Review.
Cited by
-
Tight Junction Claudins and Occludin Are Differentially Regulated and Expressed in Genomically Defined Subsets of Colon Cancer.Curr Issues Mol Biol. 2023 Oct 28;45(11):8670-8686. doi: 10.3390/cimb45110545. Curr Issues Mol Biol. 2023. PMID: 37998722 Free PMC article.
-
Human Cytomegalovirus Infection and Breast Cancer: A Literature Review of Clinical and Experimental Data.Biology (Basel). 2025 Feb 8;14(2):174. doi: 10.3390/biology14020174. Biology (Basel). 2025. PMID: 40001942 Free PMC article. Review.
-
Aberrant Expression of Claudins in Head and Neck Carcinomas and Their Prognostic and Therapeutic Value: A Narrative Review.Cancers (Basel). 2023 Aug 22;15(17):4208. doi: 10.3390/cancers15174208. Cancers (Basel). 2023. PMID: 37686483 Free PMC article. Review.
References
-
- Lehmann B.D., Jovanović B., Chen X., Estrada M.V., Johnson K.N., Shyr Y., Moses H.L., Sanders M.E., Pietenpol J.A. Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection. PLoS ONE. 2016;11:e0157368. doi: 10.1371/journal.pone.0157368. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

