SARS-CoV-2 infection alters mitochondrial and cytoskeletal function in human respiratory epithelial cells mediated by expression of spike protein
- PMID: 37504520
- PMCID: PMC10470579
- DOI: 10.1128/mbio.00820-23
SARS-CoV-2 infection alters mitochondrial and cytoskeletal function in human respiratory epithelial cells mediated by expression of spike protein
Abstract
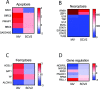
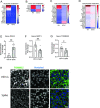
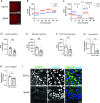
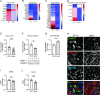
Coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2, SCV2), which has resulted in higher morbidity and mortality rate than other respiratory viral infections, such as Influenza A virus (IAV) infection. Investigating the molecular mechanisms of SCV2-host infection vs IAV is vital in exploring antiviral drug targets against SCV2. We assessed differential gene expression in human nasal cells upon SCV2 or IAV infection using RNA sequencing. Compared to IAV, we observed alterations in both metabolic and cytoskeletal pathways suggestive of epithelial remodeling in the SCV2-infected cells, reminiscent of pathways activated as a response to chronic injury. We found that spike protein interaction with the epithelium was sufficient to instigate these epithelial responses using a SCV2 spike pseudovirus. Specifically, we found downregulation of the mitochondrial markers SIRT3 and TOMM22. Moreover, SCV2 spike infection increased extracellular acidification and decreased oxygen consumption rate in the epithelium. In addition, we observed cytoskeletal rearrangements with a reduction in the actin-severing protein cofilin-1 and an increase in polymerized actin, indicating epithelial cytoskeletal rearrangements. This study revealed distinct epithelial responses to SCV2 infection, with early mitochondrial dysfunction in the host cells and evidence of cytoskeletal remodeling that could contribute to the worsened outcome in COVID-19 patients compared to IAV patients. These changes in cell structure and energetics could contribute to cellular resilience early during infection, allowing for prolonged cell survival and potentially paving the way for more chronic symptoms. IMPORTANCE COVID-19 has caused a global pandemic affecting millions of people worldwide, resulting in a higher mortality rate and concerns of more persistent symptoms compared to influenza A. To study this, we compare lung epithelial responses to both viruses. Interestingly, we found that in response to SARS-CoV-2 infection, the cellular energetics changed and there were cell structural rearrangements. These changes in cell structure could lead to prolonged epithelial cell survival, even in the face of not working well, potentially contributing to the development of chronic symptoms. In summary, these findings represent strategies utilized by the cell to survive the infection but result in a fundamental shift in the epithelial phenotype, with potential long-term consequences, which could set the stage for the development of chronic lung disease or long COVID-19.
Keywords: COVID-19; SARS-CoV-2; actin; bioenergetics; cytoskeleton; lung epithelia.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Guan W-J, Ni Z-Y, Hu Y, Liang W-H, Ou C-Q, He J-X, Liu L, Shan H, Lei C-L, Hui DSC, Du B, Li L-J, Zeng G, Yuen K-Y, Chen R-C, Tang C-L, Wang T, Chen P-Y, Xiang J, Li S-Y, Wang J-L, Liang Z-J, Peng Y-X, Wei L, Liu Y, Hu Y-H, Peng P, Wang J-M, Liu J-Y, Chen Z, Li G, Zheng Z-J, Qiu S-Q, Luo J, Ye C-J, Zhu S-Y, Zhong N-S. 2020. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 382:1708–1720. doi: 10.1056/NEJMoa2002032 - DOI - PMC - PubMed
-
- Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. 2020. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395:497–506. doi: 10.1016/S0140-6736(20)30183-5 - DOI - PMC - PubMed
-
- Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C, Pöhlmann S. 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181:271–280. doi: 10.1016/j.cell.2020.02.052 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

