A Custom Regional DNA Barcode Reference Library for Lichen-Forming Fungi of the Intermountain West, USA, Increases Successful Specimen Identification
- PMID: 37504730
- PMCID: PMC10381598
- DOI: 10.3390/jof9070741
A Custom Regional DNA Barcode Reference Library for Lichen-Forming Fungi of the Intermountain West, USA, Increases Successful Specimen Identification
Abstract
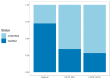
DNA barcoding approaches provide powerful tools for characterizing fungal diversity. However, DNA barcoding is limited by poor representation of species-level diversity in fungal sequence databases. Can the development of custom, regionally focused DNA reference libraries improve species-level identification rates for lichen-forming fungi? To explore this question, we created a regional ITS database for lichen-forming fungi (LFF) in the Intermountain West of the United States. The custom database comprised over 4800 sequences and represented over 600 formally described and provisional species. Lichen communities were sampled at 11 sites throughout the Intermountain West, and LFF diversity was characterized using high-throughput ITS2 amplicon sequencing. We compared the species-level identification success rates from our bulk community samples using our regional ITS database and the widely used UNITE database. The custom regional database resulted in significantly higher species-level assignments (72.3%) of candidate species than the UNITE database (28.3-34.2%). Within each site, identification of candidate species ranged from 72.3-82.1% using the custom database; and 31.5-55.4% using the UNITE database. These results highlight that developing regional databases may accelerate a wide range of LFF research by improving our ability to characterize species-level diversity using DNA barcoding.
Keywords: Illumina; OTUs; UNITE; internal transcribed spacer (ITS); metabarcoding; species hypothesis; taxonomic assignment.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Naciri Y., Linder H.P. Species delimitation and relationships: The dance of the seven veils. TAXON. 2015;64:3–16. doi: 10.12705/641.24. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

