Condensate cooperativity underlies transgenerational gene silencing
- PMID: 37505984
- PMCID: PMC10540246
- DOI: 10.1016/j.celrep.2023.112859
Condensate cooperativity underlies transgenerational gene silencing
Abstract
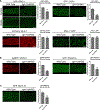
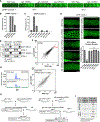
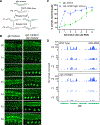
Biomolecular condensates have been shown to interact in vivo, yet it is unclear whether these interactions are functionally meaningful. Here, we demonstrate that cooperativity between two distinct condensates-germ granules and P bodies-is required for transgenerational gene silencing in C. elegans. We find that P bodies form a coating around perinuclear germ granules and that P body components CGH-1/DDX6 and CAR-1/LSM14 are required for germ granules to organize into sub-compartments and concentrate small RNA silencing factors. Functionally, while the P body mutant cgh-1 is competent to initially trigger gene silencing, it is unable to propagate the silencing to subsequent generations. Mechanistically, we trace this loss of transgenerational silencing to defects in amplifying secondary small RNAs and the stability of WAGO-4 Argonaute, both known carriers of gene silencing memories. Together, these data reveal that cooperation between condensates results in an emergent capability of germ cells to establish heritable memory.
Keywords: Argonaute; CGH-1; CP: Genomics; CP: Molecular biology; P body; PIWI; condensate; germ cells; germ granule; phase separation; small RNA; transgenerational inheritance.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




Similar articles
-
Spatiotemporal regulation of liquid-like condensates in epigenetic inheritance.Nature. 2018 May;557(7707):679-683. doi: 10.1038/s41586-018-0132-0. Epub 2018 May 16. Nature. 2018. PMID: 29769721 Free PMC article.
-
Germ Granules Coordinate RNA-Based Epigenetic Inheritance Pathways.Dev Cell. 2019 Sep 23;50(6):704-715.e4. doi: 10.1016/j.devcel.2019.07.025. Epub 2019 Aug 8. Dev Cell. 2019. PMID: 31402284 Free PMC article.
-
DEPS-1 is required for piRNA-dependent silencing and PIWI condensate organisation in Caenorhabditis elegans.Nat Commun. 2020 Aug 25;11(1):4242. doi: 10.1038/s41467-020-18089-1. Nat Commun. 2020. PMID: 32843637 Free PMC article.
-
Nuage condensates: accelerators or circuit breakers for sRNA silencing pathways?RNA. 2022 Jan;28(1):58-66. doi: 10.1261/rna.079003.121. Epub 2021 Nov 12. RNA. 2022. PMID: 34772788 Free PMC article. Review.
-
Connecting the Dots: Linking Caenorhabditis elegans Small RNA Pathways and Germ Granules.Trends Cell Biol. 2021 May;31(5):387-401. doi: 10.1016/j.tcb.2020.12.012. Epub 2021 Jan 29. Trends Cell Biol. 2021. PMID: 33526340 Review.
Cited by
-
Germ granule-mediated mRNA storage and translational control.RNA Biol. 2025 Dec;22(1):1-11. doi: 10.1080/15476286.2025.2462276. Epub 2025 Feb 6. RNA Biol. 2025. PMID: 39895378 Free PMC article. Review.
-
Nuclear Argonaute protein NRDE-3 switches small RNA partners during embryogenesis to mediate temporal-specific gene regulatory activity.bioRxiv [Preprint]. 2025 Jan 17:2024.07.29.605686. doi: 10.1101/2024.07.29.605686. bioRxiv. 2025. Update in: Elife. 2025 Mar 13;13:RP102226. doi: 10.7554/eLife.102226. PMID: 39131395 Free PMC article. Updated. Preprint.
-
Biomolecular phase separation in tumorigenesis: from aberrant condensates to therapeutic vulnerabilities.Mol Cancer. 2025 Aug 23;24(1):220. doi: 10.1186/s12943-025-02428-1. Mol Cancer. 2025. PMID: 40849651 Free PMC article. Review.
-
Nucleoporins shape germ granule architecture and balance small RNA silencing pathways.Nat Commun. 2025 May 8;16(1):4295. doi: 10.1038/s41467-025-59526-3. Nat Commun. 2025. PMID: 40341687 Free PMC article.
-
Membraneless organelles in health and disease: exploring the molecular basis, physiological roles and pathological implications.Signal Transduct Target Ther. 2024 Nov 18;9(1):305. doi: 10.1038/s41392-024-02013-w. Signal Transduct Target Ther. 2024. PMID: 39551864 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

