The contributions of rare inherited and polygenic risk to ASD in multiplex families
- PMID: 37506195
- PMCID: PMC10400943
- DOI: 10.1073/pnas.2215632120
The contributions of rare inherited and polygenic risk to ASD in multiplex families
Abstract
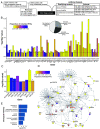
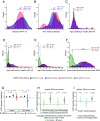
Autism spectrum disorder (ASD) has a complex genetic architecture involving contributions from both de novo and inherited variation. Few studies have been designed to address the role of rare inherited variation or its interaction with common polygenic risk in ASD. Here, we performed whole-genome sequencing of the largest cohort of multiplex families to date, consisting of 4,551 individuals in 1,004 families having two or more autistic children. Using this study design, we identify seven previously unrecognized ASD risk genes supported by a majority of rare inherited variants, finding support for a total of 74 genes in our cohort and a total of 152 genes after combined analysis with other studies. Autistic children from multiplex families demonstrate an increased burden of rare inherited protein-truncating variants in known ASD risk genes. We also find that ASD polygenic score (PGS) is overtransmitted from nonautistic parents to autistic children who also harbor rare inherited variants, consistent with combinatorial effects in the offspring, which may explain the reduced penetrance of these rare variants in parents. We also observe that in addition to social dysfunction, language delay is associated with ASD PGS overtransmission. These results are consistent with an additive complex genetic risk architecture of ASD involving rare and common variation and further suggest that language delay is a core biological feature of ASD.
Keywords: autism spectrum disorder (ASD); genetics; inherited; multiplex families; polygenic score (PGS).
Conflict of interest statement
The authors declare no competing interest.
Figures