Control of protein synthesis through mRNA pseudouridylation by dyskerin
- PMID: 37506213
- PMCID: PMC10381945
- DOI: 10.1126/sciadv.adg1805
Control of protein synthesis through mRNA pseudouridylation by dyskerin
Abstract
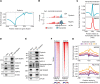
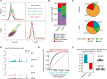
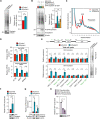
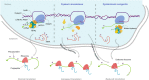
Posttranscriptional modifications of mRNA have emerged as regulators of gene expression. Although pseudouridylation is the most abundant, its biological role remains poorly understood. Here, we demonstrate that the pseudouridine synthase dyskerin associates with RNA polymerase II, binds to thousands of mRNAs, and is responsible for their pseudouridylation, an action that occurs in chromatin and does not appear to require a guide RNA with full complementarity. In cells lacking dyskerin, mRNA pseudouridylation is reduced, while at the same time, de novo protein synthesis is enhanced, indicating that this modification interferes with translation. Accordingly, mRNAs with fewer pseudouridines due to knockdown of dyskerin are translated more efficiently. Moreover, mRNA pseudouridylation is severely reduced in patients with dyskeratosis congenita caused by inherited mutations in the gene encoding dyskerin (i.e., DKC1). Our findings demonstrate that pseudouridylation by dyskerin modulates mRNA translatability, with important implications for both normal development and disease.
Figures



References
-
- F. F. Davis, F. W. Allen, Ribonucleic acids from yeast which contain a fifth nucleotide. J. Biol. Chem. 227, 907–915 (1957). - PubMed
-
- W. E. Cohn, E. Volkin, Nucleoside-5′-phosphates from ribonucleic acid. Nature 167, 483–484 (1951).
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

