Unveiling the functional epitopes of cobra venom cytotoxin by immunoinformatics and epitope-omic analyses
- PMID: 37507457
- PMCID: PMC10382524
- DOI: 10.1038/s41598-023-39222-2
Unveiling the functional epitopes of cobra venom cytotoxin by immunoinformatics and epitope-omic analyses
Abstract
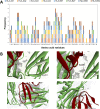
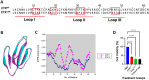
Approximate 70% of cobra venom is composed of cytotoxin (CTX), which is responsible for the dermonecrotic symptoms of cobra envenomation. However, CTX is generally low in immunogenicity, and the antivenom is ineffective in attenuating its in vivo toxicity. Furthermore, little is known about its epitope properties for empirical antivenom therapy. This study aimed to determine the epitope sequences of CTX using the immunoinformatic analyses and epitope-omics profiling. A conserved CTX was used in this study to determine its T-cell and B-cell epitope sequences using immunoinformatic tools and molecular docking simulation with different Human Leukocyte Antigens (HLAs). The potential T-cell and B-cell epitopes were 'KLVPLFY,' 'CPAGKNLCY,' 'MFMVSTPTK,' and 'DVCPKNSLL.' Molecular docking simulations disclosed that the HLA-B62 supertype exhibited the greatest binding affinity towards cobra venom cytotoxin. The namely L7, G18, K19, N20, M25, K33, V43, C44, K46, N47, and S48 of CTX exhibited prominent intermolecular interactions with HLA-B62. The multi-enzymatic-limited-digestion/liquid chromatography-mass spectrometry (MELD/LC-MS) also revealed three potential epitope sequences as 'LVPLFYK,' 'MFMVS,' and 'TVPVKR'. From different epitope mapping approaches, we concluded four potential epitope sites of CTX as 'KLVPLFYK', 'AGKNL', 'MFMVSTPKVPV' and 'DVCPKNSLL'. Site-directed mutagenesis of these epitopes confirmed their locations at the functional loops of CTX. These epitope sequences are crucial to CTX's structural folding and cytotoxicity. The results concluded the epitopes that resided within the functional loops constituted potential targets to fabricate synthetic epitopes for CTX-targeted antivenom production.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Ismail, M., al-Bekairi, A. M., el-Bedaiwy, A. M. & Abd-el Salam, M. A. The ocular effects of spitting cobras: II. Evidence that cardiotoxins are responsible for the corneal opacification syndrome. J. Toxicol. Clin. Toxicol. 31, 45–62 (1993). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

