StackTTCA: a stacking ensemble learning-based framework for accurate and high-throughput identification of tumor T cell antigens
- PMID: 37507654
- PMCID: PMC10386778
- DOI: 10.1186/s12859-023-05421-x
StackTTCA: a stacking ensemble learning-based framework for accurate and high-throughput identification of tumor T cell antigens
Abstract
Background: The identification of tumor T cell antigens (TTCAs) is crucial for providing insights into their functional mechanisms and utilizing their potential in anticancer vaccines development. In this context, TTCAs are highly promising. Meanwhile, experimental technologies for discovering and characterizing new TTCAs are expensive and time-consuming. Although many machine learning (ML)-based models have been proposed for identifying new TTCAs, there is still a need to develop a robust model that can achieve higher rates of accuracy and precision.
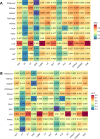
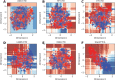
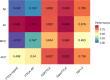
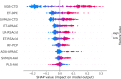
Results: In this study, we propose a new stacking ensemble learning-based framework, termed StackTTCA, for accurate and large-scale identification of TTCAs. Firstly, we constructed 156 different baseline models by using 12 different feature encoding schemes and 13 popular ML algorithms. Secondly, these baseline models were trained and employed to create a new probabilistic feature vector. Finally, the optimal probabilistic feature vector was determined based the feature selection strategy and then used for the construction of our stacked model. Comparative benchmarking experiments indicated that StackTTCA clearly outperformed several ML classifiers and the existing methods in terms of the independent test, with an accuracy of 0.932 and Matthew's correlation coefficient of 0.866.
Conclusions: In summary, the proposed stacking ensemble learning-based framework of StackTTCA could help to precisely and rapidly identify true TTCAs for follow-up experimental verification. In addition, we developed an online web server ( http://2pmlab.camt.cmu.ac.th/StackTTCA ) to maximize user convenience for high-throughput screening of novel TTCAs.
Keywords: Bioinformatics; Feature selection; Machine learning; Stacking strategy; T-cell antigen.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

