Diversity and Co-Occurrence Pattern Analysis of Cecal and Jejunal Microbiota in Two Rabbit Breeds
- PMID: 37508071
- PMCID: PMC10376057
- DOI: 10.3390/ani13142294
Diversity and Co-Occurrence Pattern Analysis of Cecal and Jejunal Microbiota in Two Rabbit Breeds
Abstract
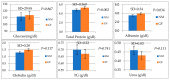
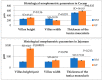
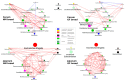
This study aimed to evaluate the productive performance and microbiota variation in the jejunum and cecum of two rabbit breeds with different growth rates. This study was carried out on Native Middle-Egypt Breed (NMER) and Giant Flanders (GF) rabbits from 5 weeks to 12 weeks of age. Twenty NMER (NM) and GF male rabbits were slaughtered, and the jejunum and cecum tracts were collected to assay gut microbiota composition via 16S ribosomal RNA (rRNA) gene sequencing and histology examination. At 12 weeks of age, daily weight gain, villus height in the jejunum, total protein, and albumin were higher in GF rabbits than in NMER rabbits. Also, the jejunal villi of GF were well arranged in their dense borders. The microbiota between the jejunum and cecum was significantly different in terms of Beta-diversity. A significant correlation between Enterococcus (jejunum NM samples) and Lactobacillus (cecum GF samples) with body weight and weight gain was found (p < 0.05). Moreover, Escherichia-Shigella in the cecum of NM was significantly correlated with weight gain (p < 0.05). The most abundant genera identified in the jejunal and cecal contents of GF were generally beneficial microbiota. They may also play a role in reducing the pathogenic effects of Escherichia coli in these rabbits.
Keywords: 16S rRNA gene sequencing; Giant Flander breed; NMER breed; breeds; cecum; growth; jejunum; microbiota; rabbits.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures





References
-
- Martinez-Guryn K., Hubert N., Frazier K., Urlass S., Musch M.W., Ojeda P., Pierre J.F., Miyoshi J., Sontag T.J., Cham C.M. Small intestine microbiota regulate host digestive and absorptive adaptive responses to dietary lipids. Cell Host Microbe. 2018;23:458–469.e455. doi: 10.1016/j.chom.2018.03.011. - DOI - PMC - PubMed
-
- Gaboriau-Routhiau V., Cerf-Bensussan N. Gut microbiota and development of the immune system. Med. Sci. M/S. 2016;32:961–967. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

