Global Antimicrobial Resistance Gene Study of Helicobacter pylori: Comparison of Detection Tools, ARG and Efflux Pump Gene Analysis, Worldwide Epidemiological Distribution, and Information Related to the Antimicrobial-Resistant Phenotype
- PMID: 37508214
- PMCID: PMC10376887
- DOI: 10.3390/antibiotics12071118
Global Antimicrobial Resistance Gene Study of Helicobacter pylori: Comparison of Detection Tools, ARG and Efflux Pump Gene Analysis, Worldwide Epidemiological Distribution, and Information Related to the Antimicrobial-Resistant Phenotype
Abstract
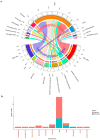
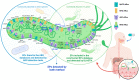
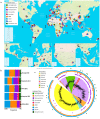
We conducted a global-scale study to identify H. pylori antimicrobial-resistant genes (ARG), address their global distribution, and understand their effect on the antimicrobial resistance (AMR) phenotypes of the clinical isolates. We identified ARG using several well-known tools against extensive bacterial ARG databases, then analyzed their correlation with clinical antibiogram data from dozens of patients across countries. This revealed that combining multiple tools and databases, followed by manual selection of ARG from the annotation results, produces more conclusive results than using a single tool or database alone. After curation, the results showed that H. pylori has 42 ARG against 11 different antibiotic classes (16 genes related to single antibiotic class resistance and 26 genes related to multidrug resistance). Further analysis revealed that H. pylori naturally harbors ARG in the core genome, called the 'Set of ARG commonly found in the Core Genome of H. pylori (ARG-CORE)', while ARG-ACC-the ARG in the accessory genome-are exclusive to particular strains. In addition, we detected 29 genes of potential efflux pump-related AMR that were mostly categorized as ARG-CORE. The ARG distribution appears to be almost similar either by geographical or H. pylori populations perspective; however, some ARG had a unique distribution since they tend to be found only in a particular region or population. Finally, we demonstrated that the presence of ARG may not directly correlate with the sensitive/resistance phenotype of clinical patient isolates but may influence the minimum inhibitory concentration phenotype.
Keywords: Helicobacter pylori; antimicrobial resistance (AMR); antimicrobial-resistant gene (ARG); clinical implications; genome analysis; global distribution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Alexander S.M., Retnakumar R.J., Chouhan D., Devi T.N.B., Dharmaseelan S., Devadas K., Thapa N., Tamang J.P., Lamtha S.C., Chattopadhyay S. Helicobacter pylori in Human Stomach: The Inconsistencies in Clinical Outcomes and the Probable Causes. Front. Microbiol. 2021;12:713955. doi: 10.3389/fmicb.2021.713955. - DOI - PMC - PubMed
-
- Hooi J.K.Y., Lai W.Y., Ng W.K., Suen M.M.Y., Underwood F.E., Tanyingoh D., Malfertheiner P., Graham D.Y., Wong V.W.S., Wu J.C.Y., et al. Global Prevalence of Helicobacter pylori Infection: Systematic Review and Meta-Analysis. Gastroenterology. 2017;153:420–429. doi: 10.1053/j.gastro.2017.04.022. - DOI - PubMed
-
- Alfaray R.I., Saruuljavkhlan B., Ansari S., Fauzia K.A., Yamaoka Y. Review: Epidemiology of Helicobacter pylori Infection. Microbiota Health Dis. 2022;4:e733. doi: 10.26355/mhd_20229_733. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

