Incidence and Genomic Background of Antibiotic Resistance in Food-Borne and Clinical Isolates of Salmonella enterica Serovar Derby from Spain
- PMID: 37508300
- PMCID: PMC10376468
- DOI: 10.3390/antibiotics12071204
Incidence and Genomic Background of Antibiotic Resistance in Food-Borne and Clinical Isolates of Salmonella enterica Serovar Derby from Spain
Abstract
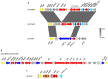
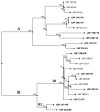
Salmonella enterica serovar Derby (S. Derby) ranks fifth among nontyphoidal Salmonella serovars causing human infections in the European Union. S. Derby isolates (36) collected between 2006 and 2018 in a Spanish region (Asturias) from human clinical samples (20) as well as from pig carcasses, pork- or pork and beef-derived products, or wild boar (16) were phenotypically characterized with regard to resistance, and 22 (12 derived from humans and 10 from food-related samples) were also subjected to whole genome sequence analysis. The sequenced isolates belonged to ST40, a common S. Derby sequence type, and were positive for SPI-23, a Salmonella pathogenicity island involved in adherence and invasion of the porcine jejune enterocytes. Isolates were either susceptible (30.6%), or resistant to one or more of the 19 antibiotics tested for (69.4%). Resistances to tetracycline [tet(A), tet(B) and tet(C)], streptomycin (aadA2), sulfonamides (sul1), nalidixic acid [gyrA (Asp87 to Asn)] and ampicillin (blaTEM-1-like) were detected, with frequencies ranging from 8.3% to 66.7%, and were higher in clinical than in food-borne isolates. The fosA7.3 gene was present in all sequenced isolates. The most common phenotype was that conferred by the tet(A), aadA2 and sul1 genes, located within identical or closely related variants of Salmonella Genomic Island 1 (SGI1), where mercury resistance genes were also present. Diverse IncI1-I(α) plasmids belonging to distinct STs provided antibiotic [blaTEM-1, tet(A) and/or tet(B)] and heavy metal resistance genes (copper and silver), while small pSC101-like plasmids carried tet(C). Regardless of their location, most resistance genes were associated with genetic elements involved in DNA mobility, including a class one integron, multiple insertion sequences and several intact or truncated transposons. By phylogenetic analysis, the isolates were distributed into two distinct clades, both including food-borne and clinical isolates. One of these clades included all SGI1-like positive isolates, which were found in both kinds of samples throughout the entire period of study. Although the frequency of S. Derby in Asturias was very low (0.5% and 3.1% of the total clinical and food isolates of S. enterica recovered along the period of study), it still represents a burden to human health linked to transmission across the food chain. The information generated in the present study can support further epidemiological surveillance aimed to control this zoonotic pathogen.
Keywords: IncI1-I(α); SGI1-like; SPI-23; ST40; Salmonella enterica serovar Derby; antimicrobial drug resistance; fosA7.3; pSC101-like; phylogenetic analysis; resistance plasmid.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Majowicz S.E., Musto J., Scallan E., Angulo F.J., Kirk M., O’Brien S.J., Jones T.F., Fazil A., Hoekstra R.M., International Collaboration on Enteric Disease ‘Burden of Illness’ Studies The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 2010;50:882–889. doi: 10.1086/650733. - DOI - PubMed
-
- WHO (World Health Organization): Salmonella (Non-Typhoidal)—Fact Sheet. 2018. [(accessed on 5 June 2023)]. Available online: http://www.who.int/mediacentre/factsheets/fs139/en/
-
- WHO (World Health Organization): Global Action Plan on Antimicrobial Resistance. World Health Organization. 2015. [(accessed on 5 June 2023)]. Available online: https://www.who.int/publications/i/item/9789241509763. - PubMed
-
- Grimont P.A.D., Weill F.X. World Health Organization Collaborating Center for Reference and Research on Salmonella. 9th ed. Institut Pasteur; Paris, France: 2007. Antigenic formulae of the Salmonella serovars.
Grants and funding
LinkOut - more resources
Full Text Sources

