Next-Generation Sequencing Technology: Current Trends and Advancements
- PMID: 37508427
- PMCID: PMC10376292
- DOI: 10.3390/biology12070997
Next-Generation Sequencing Technology: Current Trends and Advancements
Erratum in
-
Correction: Satam et al. Next-Generation Sequencing Technology: Current Trends and Advancements. Biology 2023, 12, 997.Biology (Basel). 2024 Apr 24;13(5):286. doi: 10.3390/biology13050286. Biology (Basel). 2024. PMID: 38785841 Free PMC article.
Abstract
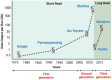
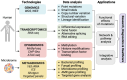
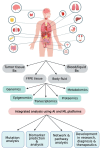
The advent of next-generation sequencing (NGS) has brought about a paradigm shift in genomics research, offering unparalleled capabilities for analyzing DNA and RNA molecules in a high-throughput and cost-effective manner. This transformative technology has swiftly propelled genomics advancements across diverse domains. NGS allows for the rapid sequencing of millions of DNA fragments simultaneously, providing comprehensive insights into genome structure, genetic variations, gene expression profiles, and epigenetic modifications. The versatility of NGS platforms has expanded the scope of genomics research, facilitating studies on rare genetic diseases, cancer genomics, microbiome analysis, infectious diseases, and population genetics. Moreover, NGS has enabled the development of targeted therapies, precision medicine approaches, and improved diagnostic methods. This review provides an insightful overview of the current trends and recent advancements in NGS technology, highlighting its potential impact on diverse areas of genomic research. Moreover, the review delves into the challenges encountered and future directions of NGS technology, including endeavors to enhance the accuracy and sensitivity of sequencing data, the development of novel algorithms for data analysis, and the pursuit of more efficient, scalable, and cost-effective solutions that lie ahead.
Keywords: Illumina; Nanopore; PacBio; bioinformatics; genomics; microbiome; molecular diagnostics; next-generation sequencing; pyrosequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources

