Targeted Protein Degradation: Principles and Applications of the Proteasome
- PMID: 37508510
- PMCID: PMC10378610
- DOI: 10.3390/cells12141846
Targeted Protein Degradation: Principles and Applications of the Proteasome
Abstract
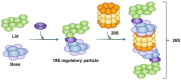
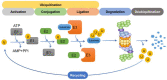
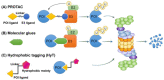
The proteasome is a multi-catalytic protease complex that is involved in protein quality control via three proteolytic activities (i.e., caspase-, trypsin-, and chymotrypsin-like activities). Most cellular proteins are selectively degraded by the proteasome via ubiquitination. Moreover, the ubiquitin-proteasome system is a critical process for maintaining protein homeostasis. Here, we briefly summarize the structure of the proteasome, its regulatory mechanisms, proteins that regulate proteasome activity, and alterations to proteasome activity found in diverse diseases, chemoresistant cells, and cancer stem cells. Finally, we describe potential therapeutic modalities that use the ubiquitin-proteasome system.
Keywords: E3 ubiquitin ligase; PROTAC; proteasome; targeted protein degradation; ubiquitin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

