Reverse-ChIP Techniques for Identifying Locus-Specific Proteomes: A Key Tool in Unlocking the Cancer Regulome
- PMID: 37508524
- PMCID: PMC10377898
- DOI: 10.3390/cells12141860
Reverse-ChIP Techniques for Identifying Locus-Specific Proteomes: A Key Tool in Unlocking the Cancer Regulome
Abstract
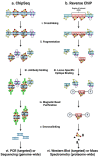
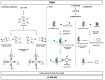
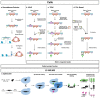
A phenotypic hallmark of cancer is aberrant transcriptional regulation. Transcriptional regulation is controlled by a complicated array of molecular factors, including the presence of transcription factors, the deposition of histone post-translational modifications, and long-range DNA interactions. Determining the molecular identity and function of these various factors is necessary to understand specific aspects of cancer biology and reveal potential therapeutic targets. Regulation of the genome by specific factors is typically studied using chromatin immunoprecipitation followed by sequencing (ChIP-Seq) that identifies genome-wide binding interactions through the use of factor-specific antibodies. A long-standing goal in many laboratories has been the development of a 'reverse-ChIP' approach to identify unknown binding partners at loci of interest. A variety of strategies have been employed to enable the selective biochemical purification of sequence-defined chromatin regions, including single-copy loci, and the subsequent analytical detection of associated proteins. This review covers mass spectrometry techniques that enable quantitative proteomics before providing a survey of approaches toward the development of strategies for the purification of sequence-specific chromatin as a 'reverse-ChIP' technique. A fully realized reverse-ChIP technique holds great potential for identifying cancer-specific targets and the development of personalized therapeutic regimens.
Keywords: cancer regulome; chromatin architecture; genome regulation; locus-specific chromatin isolation; mass spectrometry proteomics; promoter pulldown proteomics; proximity labeling; quantitative mass spectrometry; telomeres; transcriptional regulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Suzuki A., Makinoshima H., Wakaguri H., Esumi H., Sugano S., Kohno T., Tsuchihara K., Suzuki Y. Aberrant Transcriptional Regulations in Cancers: Genome, Transcriptome, and Epigenome Analysis of Lung Adenocarcinoma Cell Lines. Nucleic Acids Res. 2014;42:13557–13572. doi: 10.1093/nar/gku885. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

