Disruption of the Mammalian Ccr4-Not Complex Contributes to Transcription-Mediated Genome Instability
- PMID: 37508532
- PMCID: PMC10378556
- DOI: 10.3390/cells12141868
Disruption of the Mammalian Ccr4-Not Complex Contributes to Transcription-Mediated Genome Instability
Abstract
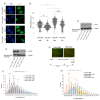
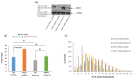
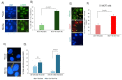
The mammalian Ccr4-Not complex, carbon catabolite repression 4 (Ccr4)-negative on TATA-less (Not), is a large, highly conserved, multifunctional assembly of proteins that acts at different cellular levels to regulate gene expression. It is involved in the control of the cell cycle, chromatin modification, activation and inhibition of transcription initiation, control of transcription elongation, RNA export, and nuclear RNA surveillance; the Ccr4-Not complex also plays a central role in the regulation of mRNA decay. Growing evidence suggests that gene transcription has a vital role in shaping the landscape of genome replication and is also a potent source of replication stress and genome instability. Here, we have examined the effects of the inactivation of the Ccr4-Not complex, via the depletion of the scaffold subunit CNOT1, on DNA replication and genome integrity in mammalian cells. In CNOT1-depleted cells, the elevated expression of the general transcription factor TATA-box binding protein (TBP) leads to increased RNA synthesis, which, together with R-loop accumulation, results in replication fork slowing, DNA damage, and senescence. Furthermore, we have shown that the stability of TBP mRNA increases in the absence of CNOT1, which may explain its elevated protein expression in CNOT1-depleted cells. Finally, we have shown the activation of mitogen-activated protein kinase signalling as evidenced by ERK1/2 phosphorylation in the absence of CNOT1, which may be responsible for the observed cell cycle arrest at the border of G1/S.
Keywords: CNOT complex; CNOT1; CNOT7; CNOT8; DNA repair; genome instability; transcription.
Conflict of interest statement
The authors have no conflict of interest.
Figures










References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

