Pyrroline-5-Carboxylate Reductase-2 Promotes Colorectal Carcinogenesis by Modulating Microtubule-Associated Serine/Threonine Kinase-like/Wnt/β-Catenin Signaling
- PMID: 37508547
- PMCID: PMC10377831
- DOI: 10.3390/cells12141883
Pyrroline-5-Carboxylate Reductase-2 Promotes Colorectal Carcinogenesis by Modulating Microtubule-Associated Serine/Threonine Kinase-like/Wnt/β-Catenin Signaling
Abstract
Background: Despite significant progress in clinical management, colorectal cancer (CRC) remains the third most common cause of cancer-related deaths. A positive association between PYCR2 (pyrroline-5-carboxylate reductase-2), a terminal enzyme of proline metabolism, and CRC aggressiveness was recently reported. However, how PYCR2 promotes colon carcinogenesis remains ill understood.
Methods: A comprehensive analysis was performed using publicly available cancer databases and CRC patient cohorts. Proteomics and biochemical evaluations were performed along with genetic manipulations and in vivo tumor growth assays to gain a mechanistic understanding.
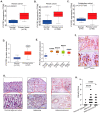
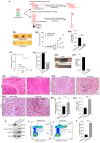
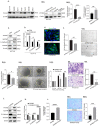
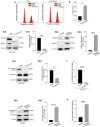
Results: PYCR2 expression was significantly upregulated in CRC and associated with poor patient survival, specifically among PYCR isoforms (PYCR1, 2, and 3). The genetic inhibition of PYCR2 inhibited the tumorigenic abilities of CRC cells and in vivo tumor growth. Coinciding with these observations was a significant decrease in cellular proline content. PYCR2 overexpression promoted the tumorigenic abilities of CRC cells. Proteomics (LC-MS/MS) analysis further demonstrated that PYCR2 loss of expression in CRC cells inhibits survival and cell cycle pathways. A subsequent biochemical analysis supported the causal role of PYCR2 in regulating CRC cell survival and the cell cycle, potentially by regulating the expression of MASTL, a cell-cycle-regulating protein upregulated in CRC. Further studies revealed that PYCR2 regulates Wnt/β-catenin-signaling in manners dependent on the expression of MASTL and the cancer stem cell niche.
Conclusions: PYCR2 promotes MASTL/Wnt/β-catenin signaling that, in turn, promotes cancer stem cell populations and, thus, colon carcinogenesis. Taken together, our data highlight the significance of PYCR2 as a novel therapeutic target for effectively treating aggressive colon cancer.
Keywords: MASTL; Wnt signaling; cancer progression; colorectal cancer (CRC); proline metabolism; proteomics; pyrroline 5 carboxylate reductases (PYCRs).
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Adorno-Cruz V., Kibria G., Liu X., Doherty M., Junk D.J., Guan D., Hubert C., Venere M., Mulkearns-Hubert E., Sinyuk M., et al. Cancer stem cells: Targeting the roots of cancer, seeds of metastasis, and sources of therapy resistance. Cancer Res. 2015;75:924–929. doi: 10.1158/0008-5472.CAN-14-3225. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

