Evaluation of Mutual Information and Feature Selection for SARS-CoV-2 Respiratory Infection
- PMID: 37508907
- PMCID: PMC10376564
- DOI: 10.3390/bioengineering10070880
Evaluation of Mutual Information and Feature Selection for SARS-CoV-2 Respiratory Infection
Abstract
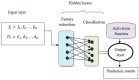
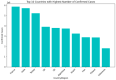
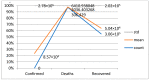
This study aims to develop a predictive model for SARS-CoV-2 using machine-learning techniques and to explore various feature selection methods to enhance the accuracy of predictions. A precise forecast of the SARS-CoV-2 respiratory infections spread can help with efficient planning and resource allocation. The proposed model utilizes stochastic regression to capture the virus transmission's stochastic nature, considering data uncertainties. Feature selection techniques are employed to identify the most relevant and informative features contributing to prediction accuracy. Furthermore, the study explores the use of neighbor embedding and Sammon mapping algorithms to visualize high-dimensional SARS-CoV-2 respiratory infection data in a lower-dimensional space, enabling better interpretation and understanding of the underlying patterns. The application of machine-learning techniques for predicting SARS-CoV-2 respiratory infections, the use of statistical measures in healthcare, including confirmed cases, deaths, and recoveries, and an analysis of country-wise dynamics of the pandemic using machine-learning models are used. Our analysis involves the performance of various algorithms, including neural networks (NN), decision trees (DT), random forests (RF), the Adam optimizer (AD), hyperparameters (HP), stochastic regression (SR), neighbor embedding (NE), and Sammon mapping (SM). A pre-processed and feature-extracted SARS-CoV-2 respiratory infection dataset is combined with ADHPSRNESM to form a new orchestration in the proposed model for a perfect prediction to increase the precision of accuracy. The findings of this research can contribute to public health efforts by enabling policymakers and healthcare professionals to make informed decisions based on accurate predictions, ultimately aiding in managing and controlling the SARS-CoV-2 pandemic.
Keywords: SARS-CoV-2 prediction; feature selection; machine learning; neighbor embedding; sammon mapping; stochastic regression.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Bhattacharya M., Alshammari A., Alharbi M., Dhama K., Lee S.S., Chakraborty C. A novel mutation-proof, next-generation vaccine to fight against upcoming SARS-CoV-2 variants and subvariants, designed through AI enabled approaches and tools, along with the machine learning based immune simulation: A vaccine breakthrough. Int. J. Biol. Macromol. 2023;17:124893. doi: 10.1016/j.ijbiomac.2023.124893. - DOI - PMC - PubMed
-
- Moitra P., Chaichi A., Hasan S.M., Dighe K., Alafeef M., Prasad A., Gartia M.R., Pan D. Probing the mutation independent interaction of DNA probes with SARS-CoV-2 variants through a combination of surface-enhanced Raman scattering and machine learning. Biosens. Bioelectron. 2022;208:114200. doi: 10.1016/j.bios.2022.114200. - DOI - PMC - PubMed
-
- Beduk D., de Oliveira Filho J.I., Beduk T., Harmanci D., Zihnioglu F., Cicek C., Sertoz R., Arda B., Goksel T., Turhan K., et al. ‘All In One’ SARS-CoV-2 variant recognition platform: Machine learning-enabled point of care diagnostics. Biosens. Bioelectron. X. 2022;10:100105. doi: 10.1016/j.biosx.2022.100105. - DOI - PMC - PubMed
-
- Taft J.M., Weber C.R., Gao B., Ehling R.A., Han J., Frei L., Metcalfe S.W., Overath M.D., Yermanos A., Kelton W., et al. Deep mutational learning predicts ACE2 binding and antibody escape to combinatorial mutations in the SARS-CoV-2 receptor-binding domain. Cell. 2022;185:4008–4022. doi: 10.1016/j.cell.2022.08.024. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

