Assessment of Colorectal Cancer Risk Factors through the Application of Network-Based Approaches in a Racially Diverse Cohort of Colon Organoid Stem Cells
- PMID: 37509213
- PMCID: PMC10377524
- DOI: 10.3390/cancers15143550
Assessment of Colorectal Cancer Risk Factors through the Application of Network-Based Approaches in a Racially Diverse Cohort of Colon Organoid Stem Cells
Abstract
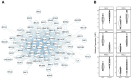
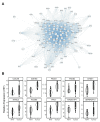
Numerous demographic factors have been associated with colorectal cancer (CRC) risk. To better define biological mechanisms underlying these associations, we performed RNA sequencing of stem-cell-enriched organoids derived from the healthy colons of seven European Americans and eight African Americans. A weighted gene co-expression network analysis was performed following RNA sequencing. Module-trait relationships were determined through the association testing of each module and five CRC risk factors (age, body mass index, sex, smoking history, and race). Only modules that displayed a significantly positive correlation for gene significance and module membership were considered for further investigation. In total, 16 modules were associated with known CRC risk factors (p < 0.05). To contextualize the role of risk modules in CRC, publicly available RNA-sequencing data from TCGA-COAD were downloaded and re-analyzed. Differentially expressed genes identified between tumors and matched normal-adjacent tissue were overlaid across each module. Loci derived from CRC genome-wide association studies were additionally overlaid across modules to identify robust putative targets of risk. Among them, MYBL2 and RXRA represented strong plausible drivers through which cigarette smoking and BMI potentially modulated CRC risk, respectively. In summary, our findings highlight the potential of the colon organoid system in identifying novel CRC risk mechanisms in an ancestrally diverse and cellularly relevant population.
Keywords: WGCNA; aging; body mass index; colon organoid; colorectal cancer risk; network analysis; racial disparities; smoking; stem cell.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures



Similar articles
-
Novel insights into the molecular mechanisms underlying risk of colorectal cancer from smoking and red/processed meat carcinogens by modeling exposure in normal colon organoids.Oncotarget. 2021 Sep 14;12(19):1863-1877. doi: 10.18632/oncotarget.28058. eCollection 2021 Sep 14. Oncotarget. 2021. PMID: 34548904 Free PMC article.
-
Potential role of fructose on human colon DNA methylation in racial disparities observed for colorectal cancer risk.medRxiv [Preprint]. 2023 Jun 4:2023.05.31.23290777. doi: 10.1101/2023.05.31.23290777. medRxiv. 2023. Update in: Am J Clin Nutr. 2025 Mar;121(3):522-534. doi: 10.1016/j.ajcnut.2025.01.005. PMID: 37398462 Free PMC article. Updated. Preprint.
-
Identification of key genes involved in the development and progression of early-onset colorectal cancer by co-expression network analysis.Oncol Lett. 2020 Jan;19(1):177-186. doi: 10.3892/ol.2019.11073. Epub 2019 Nov 8. Oncol Lett. 2020. PMID: 31897128 Free PMC article.
-
Single-cell transcriptomics reveals stage- and side-specificity of gene modules in colorectal cancer.Res Sq [Preprint]. 2024 May 24:rs.3.rs-4402565. doi: 10.21203/rs.3.rs-4402565/v1. Res Sq. 2024. PMID: 38826219 Free PMC article. Preprint.
-
Holistic exploration of CHGA and hsa-miR-137 in colorectal cancer via multi-omic data Integration.Heliyon. 2024 Mar 3;10(5):e27046. doi: 10.1016/j.heliyon.2024.e27046. eCollection 2024 Mar 15. Heliyon. 2024. PMID: 38495181 Free PMC article.
Cited by
-
Hsa_circ_0004194 suppresses colorectal cancer progression via hsa-miR-27a-3p.Heliyon. 2024 Oct 20;10(20):e39549. doi: 10.1016/j.heliyon.2024.e39549. eCollection 2024 Oct 30. Heliyon. 2024. PMID: 39498085 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

