Landscape of Genetic Mutations in Appendiceal Cancers
- PMID: 37509254
- PMCID: PMC10377024
- DOI: 10.3390/cancers15143591
Landscape of Genetic Mutations in Appendiceal Cancers
Abstract
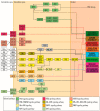
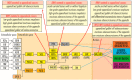
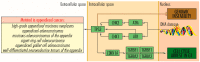
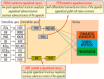
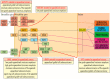
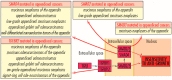
In appendiceal cancers, the most frequently mutated genes are (i) KRAS, which, when reactivated, restores signal transduction via the RAS-RAF-MEK-ERK signaling pathway and stimulates cell proliferation in the early stages of tumor transformation, and then angiogenesis; (ii) TP53, whose inactivation leads to the inhibition of programmed cell death; (iii) GNAS, which, when reactivated, links the cAMP pathway to the RAS-RAF-MEK-ERK signaling pathway, stimulating cell proliferation and angiogenesis; (iv) SMAD4, exhibiting typical tumor-suppressive activity, blocking the transmission of oncogenic TGFB signals via the SMAD2/SMAD3 heterodimer; and (v) BRAF, which is part of the RAS-RAF-MEK-ERK signaling pathway. Diverse mutations are reported in other genes, which are part of secondary or less critical signaling pathways for tumor progression, but which amplify the phenotypic diversity of appendiceal cancers. In this review, we will present the main genetic mutations involved in appendix tumors and their roles in cell proliferation and survival, and in tumor invasiveness, angiogenesis, and acquired resistance to anti-growth signals.
Keywords: KRAS genes; adenocarcinomas; appendix tumor; point mutations; signaling pathways.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- De Souza S.C., da Costa S.R.M.R., de Souza I.G.S. Vermiform appendix: Positions and length—A study of 377 cases and literature review. J. Coloproctol. 2015;35:212–216. doi: 10.1016/j.jcol.2015.08.003. - DOI
-
- Amin M.B., Greene F.L., Edge S.B., Compton C.C., Gershenwald J.E., Brookland R.K., Meyer L., Gress D.M., Byrd D.R., Winchester D.P. Appendix: Carcinoma. In: Amin M.B., editor. AJCC Cancer Staging Manual. 8th ed. Springer; New York, NY, USA: 2017. - PubMed
-
- Nagtegaal I.D., Klimstra D.S., Washington M.K. WHO Classification of Tumors: Digestive System Tumors. 5th ed. International Agency for Research on Cancer; Lyon, France: 2019. Tumors of the appendix.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

